Poaceae (Angiosperm Phylogeny)
Extracted from Angiosperm Phylogeny Website
http://www.mobot.org/mobot/research/apweb/welcome.html
By P.F. Stevens and E.A. Kellogg
- Poaceae can be recognised by their leaves that have long, usually open
sheaths and ligules at the sheath-lamina junction, round and often hollow stems,
and inflorescences whose basic unit is a spikelet. Individual flowers are small,
lacking an obvious perianth and with a gynoecium that usually has two plumose
stigmas and a single ovule. In the dry, achenial fruit (often called a
caryopsis), the rather large embryo is lateral, lying next to the seed
coat.
- The large tribe Andropogoneae, with almost 1,100 species, can be
distinguished from all other grasses because they have paired
spikelets.
Evolution. Stem-group Poaceae are dated to
ca 89 million years before present, the crown group diverge ca 83 million years
before present (Janssen & Bremer 2004: Streptochaeta included, see
also Bremer 2002; dates in Wikström et al. 2001 are far younger).
Bouchenak-Khelladi et al. (2009, 2010a) suggest that grasses originated ca 90
million years ago, although Bouchenak-Khelladi et al. (2010c) estimated that the
basal split in the family was rather younger, (86-)68(-53) million years, i.e.
in the Late Cretaceous (in their Fig. 1 it is ca 72 million years).
Interestingly, Bouchenak-Khelladi et al. (2010c) suggest that the family
originated in Africa; Bremer () had suggested that its origin was in South
America - either way, it seems to be Gondwanan. The family may initially have
been forest dwellers, and now species-poor clades, as well as the stem PACCMAD
clade, may have diverged by the end of the Cretaceous (Bouchenak-Khelladi et al.
2010c - but Puelioideae not included).
However, Poinar (2004) proposed that Programinis
burmitis, fossil in the Early Cretaceous of Myanmar some 100-110 million
years before present, was an early bambusoid grass type. Although it has some
vegetative features that are common in Poaceae, it does not have distinctive
features of the family and so is unlikely to be included here (Caroline
Stömberg, pers. comm.; Smith et al. 2010). The age of grasses (as well as that
of other monocot groups, not to mention the animals, both vertebrates and
insects, associated with them) is also questioned by the discovery of
well-preserved phytoliths of types to be found in the PACCMAD and BEP clades in
coprolites of sauropod dinosaurs from the Late Cretaceous (71-65 million years
before present) of central India (Prasad et al. 2005: there are grass pollen and
macrofossils also known from this age), and this would date the origination of
the clade to some 85-80 million years ago. This record, too, needs confirmation,
although the enigmatic Late Cretaceous mammalian sudamericid gondwanatherians
also had hypsodont teeth and there is a record of a hadrosaurian dinosaur with
carbon isotope ratios that suggests that it might have been eating C4
plants (Prasad et al. 2005; Bocherens et al. 1993). The bottom line is that
dates from these different lines of evidence are apparently irreconcilably in
conflict (Vicentini et al. 2008).
As to more conventional grasses (the [PACCMAD +
BEP] clade), fossil spikelets assignable to them are known from the
Palaeocene-Eocene boundary, about 55 million years before present (Crepet &
Feldman 1991), and this estimate is broadly in line with an estimate of the age
of a genome duplication in Poaceae (70-50 million years before present: Blanc
& Wolfe 2004; Schlueter et al. 2004; Paterson et al. 2004; Kim et al. 2009)
and other estimates like that of Vicentini et al. (2008) and Bouchenak-Khelladi
et al. (2010a) - (60-)52(-44) million years old. Bouchenak-Khelladi et al.
(2009, 2010a, c) suggested that the BEP clade began to diversify about 55-35
million years ago and the PACCMAD clade rather later some 45-37 million years
ago (see Bouchenak-Khelladi et al. 2010a for other dates; Bouchenak-Khelladi et
al. 2010c suggest (55-)52(-50) and (34-)28(-22) million years respectively).
Bouchenak-Khelladi et al. (2009, see also 2010a, c) suggested that Bambusoideae
diversified only some (39-)32(-24) million years ago in the middle
Oligocene.
The Poaceae group of families has been described as
being notably speciose (Magallón & Sanderson 2001), but there is
considerable asymmetry in family size within this clade, with most
species belonging to Poaceae themselves. The second most species-rich family
(Restionaceae) has only some 520 species. Poaceae themselves may be seven times
more speciose than its animal-pollinated sister clade (Kay & Sargent 2009).
But again, even within Poaceae there are three species-poor clades that are
successively immediately sister below the PACCMAD and BEP clades (there are two
more such clades successively sister below the family), so calling the whole
family speciose is an overstatement. If there is a focus on diversification,
this may have occurred within the PACCMAD and BEP clades (see also Linder &
Rudall 2005; Smith et al. 2011; and especially Bouchenak-Khelladi et al. 2010c
for diversification). Such statements can be refined, as by Bouchenak-Khelladi
et al. (2010c).
Hodkinson et al. (2008) discuss increases of
diversification rates in Poaceae in the context of a supertree; there seems to
have been one increase when true spikelets developed, and several others
elsewhere in the family (see also Bouchenak-Khelladi et al. 2010c). The
herbaceous habit and annual life cycle appear to be correlated with species
richness (Salamin & Davies 2004; Smith & Donoghue 2008).
Much has been written on the evolution of
C4 photosynthesis in grasses, e.g. see Kellogg (1999), Giussani et
al. (2001: Paniceae), etc. C4 photosynthesis may have evolved up to
eight times in Panicoideae alone (Giussani et al. 2001); it has also evolved
independently in Micrairoideae, Aristidoideae (twice) and Chloridoideae, for a
total of perhaps ca 18 independent origins (Kellogg 2000 and references;
Christin et al. 2008, 2009a, b; Vicentini et al. 2008; Cerros-Tlatilpa &
Columbus 2009 and Christin & Besnard 2009 [both Aristidoideae]; Sage et al.
2011). Details of the mechanisms of C4 photosynthesis and the
morphologies associated with it are very variable in Panicoideae, and
C4 photosynthesis has evolved more than once there (Kellogg 2000 and
references; see also Giussani et al. 2001; Christin et al. 2007a, 2009a).
Furthermore, taxa like Miscanthus x giganteus carry out this kind of
photosynthesis under decidedly cooler conditions than is common (Wang et al.
2008). It has been suggested that the relatively uncommon C4 PCK
subtype (phosphoenolpyruvate carboxykinase) is basal in Chloridoideae,
being subsequently lost and reacquired (Christin et al. 2009b, but see Christin
et al. 2010a for reversals; Ingram et al. 2011b for a reversal that wasn't).
Indeed, the level of parallelisms here may be at the amino acid, similar changes
occurring independently in the phosphoenolpyruvate carboxylase gene in
grasses (Christin et al. 2007a, esp. b, 2009a), in particular, a mutation to
serine at position 780 seems to have occurred in all plants with C4
photosynthesis (Bläsing et al. 2000; see also Brown et al. 2011 for parallelisms
between grasses and Capparidaceae). The adoption of C4 photosynthesis
is associated with well-known anatomical changes, such as closer spacing of the
veins, the development of a sheath of chloroplast-rich cells around the vascular
bundles, etc., and anatomy is also used to characterize subtypes of
photosynthetic pathways, however, the correlation may not be that good (Ingram
2010), and the typology needs to be revisited (E. A. Kellogg, pers. comm.).
In warmer grasslands C4 grasses now
predominate, and all told C4 photosynthesis accounts for about 18-21%
of terrestrial gross primary productivity (Lloyd & Farquhar 1994; Ehleringer
et al. 1997), while it has been suggested that grasslands - both C3
and C4 species are of course involved - currently account for 11-19%
of net primary productivity on land and 10-30% of soil C storage (Hall et al.
2000). Over half the species with the C4 photosynthetic pathway occur
in Poaceae - in total there are a mere 6,000-6,500 species involved (R. F. Sage,
pers. comm.) of which probably somewhat under 4,600 species are grasses (Sage et
al. 1999).
The balance of the evidence suggests that
C4 photosynthesis in grasses appeared first in the Oligocene, some 32
million years ago as global CO2 levels in the atmosphere declined
(e.g. Christin et al. 2008, 2011b; Vicentini et al. 2008; Bouchenak-Khelladi et
al. 2009 - but see below). It is known from grasses from the Early to Middle
Miocene in both the Great Plains and Africa, some 25-12.5 million years before
present, as C4 photosynthesis became energetically advantageous in
some environments (e.g. Ehleringer 1997 and references; Christin et al. 2008,
2011b). The great expansion of grasses with C4 photosynthesis seems
to have occurred considerably later - i.e. in the late Miocene only 9-4 million
years before present - than the initial evolution of this photosynthetic
syndrome. However, why C4 grasses spread in the way they did is still
not well understood. Increasing temperature, open habitats, and perhaps
especially decreasing precipitation (e.g. Edwards & Still 2007; Edwards et
al. 2007; esp. Edwards & Still 2008 - although by no means all C4
grasses are drought tolerant; Edwards 2009), the high flammability of dry
grasses, and windiness are additional factors that would lead to the increased
occurrence of fires, which would have removed trees - and also nitrogen by
volatilizing it - from some habitats and hence favoured grasses, C4
grasses have a reduced requirement for photosynthetic enzymes and so a lower
nitrogen requirement (Wedin 1995). The relatively low nitrogen content in grass
litter means that this builds up, so making grasslands more susceptibe to fire
(Wedin 1995). Grasses also have dense root masses that would make the
establishment of woody vegetation in grassland difficult. Interestingly, both
origins of and reversals from C4 photosynthesis may be clustered
(Vicentini et al. 2008, for reversals from C4, see also Ibrahim et
al. 2009).
Which of the various factors favoured the initial
expansion of grasslands, and what might cause clustering of origins and losses
of different photosynthetic mechanisms, and which favoured the great spread and
expansion to dominance of late Miocene C4 grasslands, is unclear (see
also Tipple & Pagani 2007; Christin et al. 2008, for the early origins of of
C4 photosynthesis and its subsequent development; Jacobs et al. 1989
and especially Retallack 2001 for the paleoecology of Poaceae; Sage & Kubien
2003; Fox & Koch 2004; Osborne & Beerling 2006; Bond 2008; Osborne 2008
for fires, etc.; Osborne & Freckleton 2009: open habitats, then drier
conditions; Arakaki et al. 2011). Recent work suggests that many C4
origins are correlated with a reduction in annual rainfall - indeed, grasslands
transpire less than the woodlands they seem iniitially to have replaced
(Retallack 2001), and this may be connected with declining late Miocene
temperatures, as Arakaki et al. (2011) point out - which perhaps favoured the
expansion of more open habitats in which these grasses thrived (Edwards &
Smith 2010). Taylor et al. (2010) and Ripley et al. (2010) make ecophysiological
comparisons between C3 and C4 grasses, the latter
sometimes being more sensitive to drought and recovering more slowly from it.
But in the late Miocene declining CO2 in the atmosphere may also have
helped things along again (Arakaki et al. 2011).
Cooler temperate grasslands are dominated by the
derived Poöideae, all of which are C3 grasses, and understanding
diversification in Poöideae entails understanding the evolution of cold
tolerance (Edwards 2009; Edwards & Smith 2010). Core Poöideae evolution may
be linked with a period of cooling at the beginning of the Oligocene ca 33-27
million years ago (Strömberg 2005; Sandve et al. 2008; Sandve & Fjellheim
2010). Gene families implicated in low temperature stress response expanded
prior to Poöideae diversification (Sandve & Fjellheim 2010), and proteins
that inhibit ice recrystallization are known from the group (Sandve et al. 2010;
Tremblay et al. 2005). There is a complex relationship between day length,
freezing tolerance and flowering (Dhillon et al. 2010). Furthermore, although
only low levels of fructan - specifically levans - accumulation have been noted
in many Poaceae, notably high levels are found only in Poöideae, although not in
the taxa of the "basal" pectinations (see Hendry 1993 for taxa involved; Pollard
& Cairns 1991). Storing carbohydrates as fructans may enable these Poöideae
to survive drought or frost better, fructans being implicated in stabilizing
cell membranes at low temperatures (Livingston et al. 2009; Sandve &
Fjellheim 2010). Another factor contributing to the diversification of Poöideae
may be the establishment of vernalization (Preston & Kellogg 2008), although
how widely this occurs outside the subfamily is unclear. Finally, the
establishment of the Epichloë/Poöideae relationship may have been
involved in the spread of Poöideae from shady to sunny habitats in the
predominantly cool-season climates that they favor (Kellogg 2001), the mutualism
aiding the plant's defences against herbivores and drought (Schardl et al. 2008;
Schardl 2010).
Grasses now cover about 20% of the land surface,
about half that area being within the tropics (Hall et al. 2000; Sabelli &
Larkins 2009 for references). Open-habitat grasses - these were mostly
C3 grasses initially - diversified taxonomically in North America in
the early Oligocene ca 34 million years ago and became ecologically dominant in
the late Oligocene to early Miocene 7-11 million years later (Strömberg 2005),
although grasslands of the Great Plains may be late Oligocene ca 24 millions
year in age and Argentinian grasslands older (Edwards et al. 2010). Some
C4 grasses may have originated in the Oligocene ca 33 million years
ago, but they became diverse - and made a corresponding major contribution to
overall vegetation biomass - only in the late Miocene-early Pliocene 9-8 million
years ago, the process being complete as recently as a mere 3-2 million years
ago (Bouchenak-Khelladi et al. 2009; Edwards et al. 2010; Strömberg &
McInerney 2011; McInerny et al. 2011 for North America; Arakaki et al. 2011).
The spread of grasslands may be associated with a CO2 decrease
(Arakaki et al. 2011) perhaps connected with the activities of ectomycorrhizal
taxa (Taylor et al. 2009; see Gerhart and Ward 2010 and Zachos et al. 2008 for
past CO2 concentrations) which made trees less competitive (Pagani et
al. 2009); grasslands seem to have expanded at the expense of woodlands
(Retallack 2001), although McInerey et al. (2011) suggest that the late Neogene
expansion of C4 grasses in North America was at the expense of
C3 grasses rather than woody vegetation. What makes grasslands still
more distinctive ecologically is not just the habit of grasses, but that
relatively few species of grasses are ecologically dominant in any one
place, and most of these seem to be C4 grasses (Edwards et al. 2010).
A perhaps ecologically related feature of a number
of grasses scattered in different subfamilies is their accumulation of glycine
betaines and other compounds commonly associated with allowing plants containing
them to grow in saline conditions (Rhodes & Hanson 1993). Some Poaceae have
allelopathic reactions with other plants, Sorghum roots producing a
quinone (an oxygen-substituted aromatic compound) and Festuca roots
meta-tyrosine, a non-protein amino acid (Bertin et al. 2007).
Benzoxazinoids, cyclic hydroxamic acids, are known from members of Poaceae
including both Panicoideae and Poöideae; they confer resistance to fungi,
insects, and even herbicides, as well as being allelopathic (Frey et al. 1997,
2009); they are very uncommon in other angiosperms (Schullehner et al. 2008).
Sindhu et al. (2008) note that the PACCMAD clade are characterized by a gene
that protects the plant against attack by the the ascomycete Cochliobolus
carbonorum. Finally, leaves of pooid monocots (presumably including sedges)
decompose more slowly than do those of other angiosperms (Cornwell et al. 2008).
There are additional eco-physiological factors to
bear in mind. The dumb-bell shaped stomata of grasses show remarably rapid
stomatal movements, very much faster than those few other stomata have been
examined (Franks & Farquhar 2006). However, the significance of this is
unclear given that quite a number of other Poales have similar stomata; whether
the evolution of these stomata is a major component of th ability of grasses to
spread as climates became drier at the end of the Eocene remains to be seen
(Hetherington " Woodward 2003). Prominent rhizosheaths - mucilage from root cap
cells, soil particles, bacteria, etc., all anchored to root hairs - occur in
many Poaceae (McCulley 1995), especially those growing in drier conditions,
although the distribution of such roots is poorly known - they certainly occut
in other Poales, but are rare in broad-leaved angiosperms. Interestingly,
C4 grasses have roots with long root hairs yet may respond positively
in terms of phosphorus uptake when forming endomycorrhizal associations - long
root hairs and endomycorrhizal associations tend to be thought of as alternative
ways of securing phosphorus supply, etc. (Schweiger et al. 1995 and references).
Finally, it may be worth mentioning that Poaceae, apparently alone in flowering
plants (Römheld 1987), acquire iron through chelation of ferric ions with
siderophores which are then taken up by the roots; iron (and zinc) are commonly
limiting trace elements in alkaline soils (Schmidt 2003; Kraemer et al. 2006).
Ectomycorrhizal plants, also noted for dominating the communities in which they
occur, also produce siderophores......
There was a Miocene radiation of grazing mammals
(Thomasson & Voorhies 1990) that may be associated with the spread of such
prairie and savannah grasses (see also Cerling et al. 1997; Bouchenak-Khelladi
et al. 2009, 2010a, the latter with considerable detail and many dates;
Mihlbachler et al. 2011). These mammals evolved hypsodont dentition, i.e. teeth
with high crowns, enamel extending below the gum lines, and short roots to deal
with the wear caused by eating the abrasive grasses (but see below - Sanson
& Heraud 2010); the persistent dead leaves of most grasses may have
decreased their palatability (Antonelli et al. 2010). C4 grasses may
be less palatable than C3 grasses, having more sclerenchyma because
the veins are closer (see Caswell et al. 1973 in part), although the nitrogen
content of C3 and C4 grasses seems to be similar (Taylor
et al. 2010). However, although prairie grasses expanded in Nebraska in the
Early Miocene ca 23 million years before present, hypsodont ungulates were
already around by then (Strömberg 2004); Bovidae and Cervidae started
diversifying at least 26 million years ago (Bouchenak-Khelladi et al. 2009).
Massive diversification of ungulates is largely a Miocene phenomenon
(Bouchenak-Khelladi et al. 2009), and specialists on C4 grasses seem
to have evolved before those grasses dominated (Edwards et al. 2010).
Bouchenak-Khelladi et al. (2009) noted that the density of silica bodies in the
leaf epidermis seems to have increased in a number of grass groups, perhaps as a
defence against herbivory, however, Sanson and Heraud (2010) suggested that the
silica there might not be in crystalline form and so be unable to cause wear on
the enamel of mammalian teeth. In New Zealand, lacking mammalian herbivores,
Danthonioideae in particular may have tended to become deciduous, increasing
their productivity (Antonelli et al. 2010). Establishing connections between the
evolution and rise to dominance of grasses in some ecosystems and the evolution
of grazing animals needs more work, but it seems to me likley that the two are
linked (see also Retallack 2001 for a good summary).
Clavicipitaceous endophytes (class 1 endophytes:
Rodriguez et al. 2009) are widely distributed among grasses. Leuchtmann (1992)
discussed the distribution and host specificity of grass endophytes in general
(Clay 1990 is a still useful general review; see also Schardl 2010). Some 30% or
more of Poöideae are involved in such associations, and both horizontal and in
particular vertical transmission of these fungi occurs. They can be placed in
ascomycetes-Clavicipitaceae-Balansiae (Clay 1986). This endophyte-grass
relationship is usually described as being one of mutualism, although this may
sometimes, at least, not be so (see Saikkonen et al. 1998; Gundel et al. 2006;
Ren & Clay 2009), and the more that is found out about this relationship,
the more complex it appears to be. One of the most important fungi involved is
Epichloë (Clavicipitaceae), a systemic endophyte restricted to Poöideae;
Neotyphodium is its asexual stage, perhaps hybrids of Epichloë
species (Roberts et al. 2005; Moon et al. 2005). For its phylogeny and evolution
see Schardl (1996, 2002, 2010), Craven et al. (2001), Clay and Schardl (2002),
Jackson (2004 [possible codivergence]), and Gentile et al. (2005), and for the
patterns of infection of the two forms, see Rodgers et al. (2009). There are
four groups of alkaloids that are synthesized by Epichloë: indole
diterpenes, lolines, peramine, and the ergot alkaloids (Fleetwood et al. 2007).
Indeed, a variety of "grass" alkaloids, including loliine (pyrrolizidine) and
ergot alkaloids (ergolines), are in fact synthesized by the fungal member of
this association, which could be ca 40 million years old (Schardl et al. 2004).
These loliine alkaloids are primarily active against insects (Schardl et al.
2007; Zhang et al. 2009), and so the presence of endophytes affects the
palatability of grasses to herbivores and of their seeds to granivorous birds,
the animals eating the infected material sometimes not thriving at all; the
level of aphid infestation and that of their parasites and parasitoids,
infestation by nematodes, and even the pattern and rate of decomposition of dead
grass are also affected (e.g. Madej & Clay 1991 - birds; Popay & Rowan
1994 - general; Omacini et al. 2001 - aphids; Lemmons et al. 2005 -
decomposition; Schardl 2010). Furthermore, the larvae of Phorbia (or
Botanophila) flies live on Epichloë stroma, and the adults
transmit the spermatia in a fashion analogous to insect pollination of flowers
(Bultman 1995), indeed, Epichloë synthesizes unique compounds that
specifically attract the flies (Steinebrunner et al. 2008) and which may also be
toxic to other fungi that secondarily invade the fungal stromata (Schiestl et
al. 2006). However, the equilibrium of such relationships can easily be
disturbed (Eaton et al. 2010).
Large numbers of other apparently symptomless
endophyte species (class 3 endophytes) may grow together on Poaceae, but little
is known about their interactions with the host. Márquez et al. (2007) found
that only when the endophytic fungus (Curvularia) was infected with a
virus was Dicanthelium lanuginosum, the host of the fungus, able to grow
in volcanically-heated soils, suggesting the complexity of such relationships,
while Marks and Clay (2007) discuss growth rate of endophyte-infected and -free
plants under various conditions. For fungal records - very numerous and diverse
- on grasses, see Tang et al. (2007); there are at least 1933 species of fungi
from bamboos alone. Some root-associated endophytic fungi (class 4) are also
coprophilic fungi (Herrera et al. 2009), perhaps aiding in their dispersal.
Bacteria may be endophytes too, and several
bacterial endophytes are implicated in fixing 1/3 to 1/5 the nitrogen needed by
sugarcane in Brazil - the bacteria include Gluconacetobacter
(alpha-Proteobacteria) and Herbaspirillum and Burkholderia
(ß-Proteobacteria, for the latter, see also Fabaceae) (de Carvalho et al.
2011).
Poaceae provide food for both adults (as pollen)
and larvae (as roots) of Chrysomelidae-Galerucinae-Luperini-Diabrotica
beetles (Jolivet & Hawkeswood 1995). Caterpillars of nymphalid butterflies,
in particular the browns, Satyrini, and the related Morphini, are common (over
10% of the records); the satyrine group as a whole diverged from other
Nymphalidae some 85 million years ago (Wahlberg et al. 2009), Satyrini
themselves diverging from the rest of the group about 65-55 million years ago
(Peña & Wahlberg 2008; Wahlberg et al. 2009 - age depends on calibration
points used, the position of Satyrini within the satyrine clade differs
greatly). Indeed, larvae of Satyrinae-Satyrini, with some 2,200 species, the
bulk of the clade, almost exclusively eat grasses, and the main lineages within
Satyrini diversified well after the initial divergence of Satyrini and only some
36–23 Myr ago - an age perhaps contemporaneous with the spread of grasses (see
above: Peña et al. 2006; Peña & Wahlberg 2008, dates from a tree where
Satyrini diverge ca 55 million years ago and are not sister to all other
Satyrinae). Galling diptera, especially Cecidomyiidae, are quite common here
(Labandeira 2005); Cecidomyiid gall midges, notably Mayatiola (M.
destructor is the Hessian fly), are quite common on Poöideae in North
America (Gagné 1989). Shoot flies (Diptera - Chloropidae) are gall formers on
monocots, especially grasses, but they are also simple herbivores and have other
life styles (de Bruyn 2005). Chinch bugs of the Hemiptera-Lygaeidae-Blissinae
have been most commonly observed on members of the PACCMAD clade, less commonly
on the BEP clade; Teracrini are also concentrated on Poaceae (Slater 1976). In
some grasses, at least, defence against herbivores is mediated by the production
of volatiles which attract nematodes (to attack Diabrotica larvae) or
parasitic wasps (to attack caterpillars: Degenhardt 2009).
Rusts and smuts are common on Poaceae, and those on
Bambusoideae, Poöideae (inc. Stipa and relatives) are particularly
distinctive (Savile 1979b); two thirds of Ustilaginales (smuts) - close to 600
species - are found on Poaceae (Kukkonen & Timonen 1979; Stoll et al. 2003).
Some seventy species of Berberis are alternate hosts (the aecial stage)
for Puccinia graminis, the black stem rust of wheat and other grain crops
in Poöideae - this species (or complex) infects some 77 genera of mostly pooid
grasses (Abbasi et al. 2005 and references). Cyclic hydroxamic acids are widely
distributed in the family and confer resistance against a variety of fungal and
insect pathogens (Frey et al. 1997).
Poaceae are of course predominantly wind-pollinated
with dangling anthers and protandrous flowers. However, insect pollination is
known from some forest-dwelling grasses, especially smaller Bambusoideae
(Soderstrom & Calderón 1971). Streptochaeta may also be animal
pollinated, since it lacks a plumose stigma and the anthers do not dangle; the
flowers are protogynous (Sajo et al. 2008). Woody bamboos are known for their
synchronized flowering (see below). Lodicules, modified members of the inner
tepal whorl, seem to be involved in the opening of the staminate or perfect
flowers; they can be absent from carpellate flowers (see Sajo et al. 2007;
Reinheimer & Kellogg 2009 for references).
The caryopsis is often described as being a
distinctive fruit type of the Poaceae; it is basically a variant of an achene.
In fact, there is quite a variety of fruit types in the family when it comes to
thinking about how dispersal is accomplished (e.g. Werker 1997). Dispersal is
quite often by animals, and although few Poaceae have true fruits as dispersal
units - an example is Alvimia (Bambusoideae) - there are other structures
attracting animals such as elaiosomes (Davidse 1987), as well as hooks and
spikes by which the diaspores attach to passing animals (Centotheca is a
good example). A number of taxa are wind-dispersed, for example,
Andropogon has long hairs on the awns, while Spinifex and a few
other genera are tumbleweeds. Awns can aid in both wind and animal dispersal;
the surface microstructure on awns - minute retrose bristles - may result in the
achene becoming "planted" in the ground (Elbaum et al. 2007; Humphreys et al.
2010b) or moving along the surface of the ground (Kulić et al. 2009).
This is by a ratchet principle similar to that which operates when you put an
entire inflorescence of Hordeum up your sleeve; the whole inflorescence
migrates up your arm and sometimes also down your back. Davidse (1987) notes a
number of taxa with "creeping diaspores" which can move using this mechanism.
Despite the apparent advantages of having an awn, this has been lost several
times in Danthonioideae, at least, perhaps in association with the adoption of
the annual habit where passive burial of seeds suffices (Humphreys et al.
2010b).
Woody bamboos are known for their tendency to
dominate the vegetation and their synchronized flowering, even when transported
thousands of miles from their native habitat. Flowering may occur only every 120
years or so, and after a rather protracted period of reproduction, the plant
dies. The fruits are used as food by humans and they also attract animals -
birds, rats, etc. - in very large numbers (Janzen 1976; Judziewicz et al. 1999).
This behaviour is also found in some herbaceous bamboos and, depending on
relationships within Bambusoideae, may even be plesiomorphic for the subfamily.
Water often congregates in the hollow stems of bamboos, and a distinctive fauna
lives there (Kitching 2000). Many bamboos are monocarpic - that is, the plants
dies after flowering - and this has profound effects both on the general
vegetation and all organisms dependent on them.
Relationships within Danthonioideae are complex,
and there seems to have been much reticulation in the past (Pirie et al. 2009),
as is notoriously the case in Triticeae (G. Petersen et al. 2006a; Mason-Gamer
2008; Sun & Komatsuda 2010), polyploidy and introgression further
complicating the picture.
There has been a duplication of the whole genome in
a clade that includes at least Zea, Oryza, Hordeum and
Sorghum, i.e. the PACMAD clade, and this duplication has been dated to
70-50/73-56 million years before present (Schlueter et al. 2004; The
International Brachypodium Initiative 2010). Soltis et al. (2009) suggest that
diversification in Poaceae may be connected with this genome duplication. It has
been suggested that diversification of the groups including the cereals occurred
ca 20 million years later (Paterson et al. 2004, but cf. The International
Brachypodium Initiative 2010). Malcomber and Kellogg (2005) suggest that there
has been duplication of LOFSEP genes within Poaceae, while the duplication of
AP1/FUL gene, apparently in stem-group Poaceae, may be involved in
the evolution of the spikelet (Preston & Kellogg 2006). In general,
developmental gene duplication and subsequent functional divergence seem to have
played a very important role in allowing the development of the baroque
diversity of inflorescences in the family (Malcomber et al. 2006; Zanis 2007).
Indeed, there has been very extensive duplication of genes - API,
AG and SEP families - but not in genes of the AP3 lineage
(Zahn et al. 2005a; see also Saski et al. 2007 for other duplications in the
family).
Salse et al. (2008, 2009a, b) and Abrouk et al.
(2010) discuss genome evolution in the family, suggesting that the base
chromosome number (x) is 5, but in the [PACCMAD + BEP] clade at least x
increased to 12 after a genome duplication (to x = 10) and two interchromosomal
translocations and fusions (to x = 12). n = 12 is still found in rice
(Oryza), for example, while x = 10 in Panicoideae. However, Hilu (2004)
suggested that the base chromosome number for the whole family might be x = 11.
Certainly one or more rounds of genome duplication have occurred, with
subsequent independent reductions in chromosome numbers (Schnable et al. 2009;
Abrouk et al. 2010 and references). Genome size varies considerably and at least
in part independently of chromosome number, both increasing and decreasing
(Caetano-Anollés 2005; Smarda et al. 2008; Schnable et al. 2009). Within
Poöideae there seems to have been independent reduction in chromosome number
from n = 12 (The International Brachypodium Initiative 2010). Overall, there has
been very substantial evolution in the genome of grasses, with genome evolution
in Triticeae (Poöideae) being particularly accelerated (Luo et al. 2009; see
also Messing & Bennetzen 2008; Salse et al. 2009a) - many Triticeae have
massive genomes in part because of changes in base chromosome number (Jakob et
al. 2004). Indeed, comparisons of expressed sequence tags and general genomes
suggest that the genomes of Poaceae are much more different from the genome of
Allium (Alliaceae, Asparagales) than is the genome of Arabidopsis
(Brassicaceae, Brassicales, rosid II) from that of Allium (Kuhl et al.
2004). There has also been substantial evolution in the chloroplast genome
(Guisinger et al. 2010 for literature), although details on where on the tree
(and so when) particular changes occurred await more extensive sampling of the
chloroplast genome in Poales and even "basal" Poaceae, and the rate of plastid
evolution may have since slowed down; these changes are placed at the level of
Poaceae as a whole, althougfh they might more correctly go at the PACCMAD/BEP
node...
Economic Importance. Wheat (mostly
Triticum aestivum - Poöideae), which provides one fifth of the calories
eaten by humans, began to be domesticated ca 10,000 years ago; see Israel
Journal of Plant Sciences 55(3-4). 2007, for an entry into the literature on
domestication, also Fuller (2007), Baum et al. (2009: haploid genomes) and
Carver (2009: general). Most domesticated forms are polyploid, and genome
plasticity in connection with this polyploidy has been implicated of the success
of the crop in cultivation (Dubcovsky & Dvorak 2007). For the domestication
of barley (Hordeum vulgare), see Fuller (2007), Pourkheirrandish and
Komatsuda (2007) and Azhaguvel and Komatsuda (2007). Sorghum and
Zea (Panicoideae) and Oryza (Ehrhartoideae) are three other
important grain genera. The domestication of maize seems to have occurred in
seasonal tropical forests in southwestern Mexico, perhaps the Balsas valley,
some 8,700 years before present (Piperno et al. 2009; Ranere et al. 2009:
summarized in Hastorf 2009); for a detailed summary of all aspects of maize
biology, see Bennetzen and Hake (2009). For a phylogeny of Oryzeae, see Guo and
Ge (2005), and for information on the complex history of domestication of rice
(Oryza spp.) - which occurred in two places, at least - see Sweeney and
McCouch (2007) and Fuller (2007). For the domestication of pearl millet
(Pennisetum glaucum), see Fuller (2007), and for that of sorghum
(Sorghum spp.), see Dillon et al. (2007). Sang (2008) notes that single
genes are involved in a number of major morphological transitions in the
domestication of grains, such as the development of non-shattering rhachises;
the genes may be quite different in unrelated species. For the domestication of
sugarcane (Saccharum officinarum) in New Guinea, see Dillon et al. (2007)
- note that Sorghum bicolor and Saccharum officinarum can be
hybridized (e.g. Nair 1999). Glémin and Bataillon (2009) take a comparative
viewpoint and look at how grasses in general have evolved under
domestication.
Chemistry, Morphology, etc. The primary cell
wall hemicellulose and pectin polysaccharides of grasses are very different from
that of other seed plants, both in overall composition and particularities of
the composition of the xyloglucans (O'Neill & York 2003); the
polysaccharides are less branched than those elsewhere (but overall sampling is
very poor). Hatfield et al. (2009) discuss acylation of lignin in grasses, and
Boerjan et al. (2003) note that grasses in particular have a variety of minor
lignin monomer units.
Poaceae have a nodal vascular plexus (Arber 1919),
but I have no idea as to its general distribution and significance. Microhair
variation in the family is extensive and of some use in delimiting major groups
(Amarasinghe & Watson 1988, 1990; Liu et al. 2010). Ligule variation is also
extensive: Anomochlooideae are sometimes described as lacking a ligule
(Judziewicz & Clark 2008, which see for other distinctive characters), or
the ligule is described as being represented by a ring of hairs... The leaf
blades of Neurolepis (Bambusoideae) may be up to 4 m long. The style is
hollow in Pharus. In addition, the anther wall consists solely of
epidermis and endothecium (i.e. it is of the Reduced type), the latter
degenerating before anthesis (Sajo et al. 2007). All in all, Pharus has
numerous distinctive features that need to be integrated with the phylogenetic
tree; see also Judziewicz and Clark (2008).
A common interpretation of the grass palea, which
is often bicarinate, has been that it is prophyllar/bracteolar in nature,
monocots commonly having bicarinate prophylls. However, in this scenario
bracteoles would probably have to be regained, since the immediate outgroups to
Poaceae lack them. For suggestions, based on early studies of gene expression,
that the palea and perhaps even lemma are calycine in nature and the lodicules
are corolline, see Ambrose et al. (2000); A-type genes are expressed in both the
palea and lemma (Whipple & Schmidt 2006). General comparative morphology
might suggest that the lemma is a bract and the palea represents two connate
tepals of the outer whorl; if the lemma is a perianth member, then loss of
bracts will be an apomorphy for all or most of Poaceae. The flowers of
Streptochaeta can be interpreted as having an outer perianth whorl of two
(adaxial) members that ultimately become the single, bicarinate palea (there are
sometimes three members in this outer whorl), and an inner perianth whorl of
three members that ultimately become the lodicules. The three stamens common in
grass flowers would then be those opposite the three members of the outer
perianth whorl (see Whipple & Schmidt 2006; Preston et al. 2009, and
Reinheimer & Kellogg 2009 for further details). Given the sister-group
relationships between Ecdeicoleaceae and Joinvilleaceae recently found by
Marchant and Briggs (2006) and the likelihood that the flowers of
Anomochloa are sui generis, the floral morphology of
Streptochaeta may be plesiomorphic in the family, or represent an
apomorphy for it. Recently Sajo et al. (2008) suggested that the flowers of
Streptochaeta could be interpreted in more or less conventional terms,
with a whorl of three rather coriaceous "bracts" being equivalent to lodicules
and two adaxial "bracts" outside this perhaps representing the palea (although
the structure interpreted as being a lemma was also adaxial...). Interestingly,
the flowers of Ecdeicolea are also notably monosymmetric, with the two
adaxial tepals of the outer whorl larger and keeled, and although this is not
directly relevant, comparable differentiation in the outer perianth whorl occurs
in Xyridaceae (q.v.); these are all likely to be parallelisms. The tepaloid
nature of the lodicules is relatively uncontroversial (see Sajo et al. 2007;
Reinheimer & Kellogg 2009 for references).
It is difficult to interpret the arrangement of the
pollen grains in the small anthers of Streptochaeta; they may be
peripheral, at least initially (Kirpes et al. 1996; Sajo et al. 2009). Some
grasses have pendulous, atropous ovules; although both crassi- and
tenuinucellate ovules are reported for grasses, Rudall et al. (2005a) suggest
that the reports of the former need confirmation. When there are three carpels,
the abaxial member is fertile (Kircher 1986). The caryopsis is often described
as being a distinctive fruit type of the Poaceae - here the testa and pericarp
are fused, basically a variant of an achene. Poaceae are noted for their well
developed, lateral embryo with a scutellum - nothing more than a
distinctively-shaped haustorial part of the cotyledon that is common in other
monocots (= the haustorial cotyledonary hyperphyll if one wants to be technical
- see Tillich 2007 for the grass embryo). The mitochondrial coxII.i3
intron has developed a moveable element-like sequence (Albrizio et al. 1994),
but there is a fair bit of variation in other monocots, too.
Transposable elements, Mutator-like elements
(MULEs), seem to have moved fairly recently by lateral transfer between rice,
East Asian bamboos, and a number of Andropogonoideae (Diao et al. 2006). For the
Hm1 resistance gene, see Sindhu et al. (2008), and for the complex
evolution of the Rp1 disease resistance gene family, see Luo et al.
(2010).
Some information is taken from Judziewicz and
Soderstrom (1989) and in particular from the Grass Phylogeny Working Group
(2001); a few small taxa remain unplaced in subfamilies there. For embryo
variation, see Reeder (1957), for non-starch soluble storage polysaccharides in
the seed and fructans in vegetative parts, see MacLeod and McCorquodale (1958)
and Meier and Reid (1982), for anatomy, see Metcalfe (1960), for the series of
inversions in the single copy region and expansion of the inverted repeats of
the chloroplast genome, see Hiratsuka et al. (1989), for C4
photosynthesis, see also Kellogg (1999), for accD gene loss, see Katayama
and Ogihara (1996), for phytoliths and their distribution, see Piperno and
Pearsall (1998), Piperno and Sues (2005) and Piperno (2006), for deletions,
etc., in the 3' end of the mat K gene, see Hilu & Alice (1999), for
loss of introns in chloroplast genome, see Daniell et al. (2008) for references,
and for a summary of genome evolution in the family, see Bennetzen (2007). For
the occurrence of ergot alkaloids, see Gröger and Floss (1998), for the
relationship between genus size, life form and polyploidy, see Hilu (2007a), for
inflorescence development, see Malcomber et al. (2006) and Reinheimer et al.
(2008: Paniceae), for floral/spikelet evolution, see Whippple and Schmidt
(2006), Yuan et al. (2009) and Thompson et al. (2009), for cell wall
composition, see Fincher (2009), for aerial branching, see Malahy and Doust
(2009), for aspects of inflorescence morphology, see Perreta et al. (2009), and
for endosperm development, see Sabelli and Larkins (2009). Leseberg and Duvall
(2009) look at plastome-level variation in Poaceae, and Morris and Duvall (2010)
discuss aspects of chloroplast genome evolutiom, focusing on Anomochloa.
See Bell and Bryan (2008) for a good general treatment of grass morphology, and
for a summary of grass systematics, see Hilu (2007b). Arber (1934) remains the
classic account of the family. There is also a useful general bibliography
on Poaceae, while Chase (1964) is a classic introduction to the family.
For general information on Bambusoideae, see Clark
(1997), Judziewicz et al. (1999), and Judziewicz and Clark (2008), for foliar
epidermis, see Yang et al. (2008a). For pollen in Chloridoideae, see Liu et al.
(2004: not much variation).
Phylogeny. For overviews of the family, see
Soreng and Davis (1998), Kellogg (2000a) and the Grass Phylogeny Working Group
(2001); the first in particular show how difficult it is to be sure of the
position on the tree of many of the characters mentioned above. Duvall et al.
(2010) provide a preliminary tree based on whole chloroplast genomes.
Relationships of the major clades within the PACCMAD and BEP clades are for the
most part unclear, indeed, the position of Poöideae (Hodkinson et al. 2007, and
references; Duvall et al. 2008a) and Ehrhartoideae (Cahoon et al. 2010, as
Oryzoideae) are also not clear in some analyses, however, Duvall et al. (2007)
found strong support for the BEP clade, albeit the taxon sampling was slight
(see also Saarela & Graham 2010; cf. Davis & Soreng 2008; Christin et
al. 2008: BEP clade paraphyletic and immediately basal to the PACCMAD clade).
Ehrhartoideae and Poöideae have weak to moderate support as sister taxa
(Bambusoideae not included: Saski et al. 2007: see also grass Phylogeny Working
Group 2001). Where exactly Streptogyneae are to be placed, whether in
Bambusoideae, Ehrhartoideae, or in a separate subfamily, is unclear (Hisamoto et
al. 2008). In a multi-gene study, Bouchenak-Khelladi et al. (2008) clarified
further relationships in the core Poales, while at the same time questioning
others. Thus Bouchenak-Khelladi et al. (2008) did not find strong evidence for
the monophyly of Anomochlooideae, Streptochaeta possibly being sister to
all other Poaceae; Micrairoideae may not be monophyletic, Isachne not
having a fixed position; there was support for a sister relationship between
Danthonioideae and Chloridoideae (see also Pirie et al. 2008); and
Streptogyna may be sister to the whole PACCMAD clade - and it lacks the
possible synapomorphies of that clade (Bouchenak-Khelladi et al. 2008). The
relationships obtained by Bouchenak-Khelladi et al. (2009) are largely those in
the account above. However, relationships in the PACCMAD clade are particularly
difficult (Saarela & Graham 2010, but sampling).
For a discussion of the relationships - close, and
perhaps even entwined - between Panicoideae and Centothecoideae, see Duvall et
al. (2008a) and especially Sánchez-Ken and Clark (2008); recent work suggests
that the two should be combined (Sánchez-Ken & Clark 2010). Panicoideae have
been much studied because of the important crops they contain, e.g. see Giussani
et al. (2001). For relationships in the Paniceae, see Zuloaga et al. (2000),
Gómez-Martínez and Culham (2000) and Morrone et al. (2010), for the bristle
clade of Paniceae, see Doust et al. (2007), and those within Panicum
itself, see Aliscioni et al. (2003) and Sede et al. (2008), within
Pennisetum, in which Cenchrus is embedded, see Donadío et al.
(2009) and Chemisquy et al. (2010), and within Setaria, see Kellogg et
al. (2009); see also Sede et al. (2009a) for two new genera. Salariato et al.
(2010) examined relationships within Melinidae, particularly fromn the point of
view of inflorescence evolution. Ng'uni et al. (2010) looked at relationships
with Sorghum. For general information on Paniceae, see Crins (1991), for
unisexuality, see Le Roux and Kellogg (1999), for inflorescence evolution, see
Doust and Kellogg (2002) and Reinheimer and Vegetti (2008), and for the
evolution of the NADP-malate dehydrogenase gene following its duplication, see
Rondeau et al. (2005). For the phylogeny of Andropogoneae, see Kellogg (2000c)
and Mathews et al. (2002), and for that of Paspalum, basically
monophyletic, see Rua et al. (2010). Finally, for more information on
relationships within Panicoideae, including those of some of its constituent
genera, see papers in Aliso 23: 503-562. 2008.
Relationships within Chloridoideae are something
like ]] (Peterson et al. 2009);
Peterson et al. (2010a) provide a detailed phylogeny of the clade.
Eragrostis and Sporobolus may be polyphyletic, while
Muhlenbergia is paraphyletic, including a number of well supported (and
with morphology, too) clades (Peterson et al. 2010b; Columbus et al. 2010). For
a morphological phylogenetic analysis of the subfamily, see Liu et al. (2005),
for other relationships, see papers in Aliso 23: 565-614. 2008.
For a phylogeny of the Pentaschistis group
(Danthonioideae), also character evolution there, see Galley & Linder
(2007), for relationships in the subfamily as a whole, see Barker et al. (2007a)
and Pirie et al. (2008). Some relationships within Danthonioideae are
reticulating (Pirie et al. 2009).
Zhang and Clark (2000) clarified relationships of
Bambusoideae, restricting the limits of the subfamily to that now generally
accepted; most of the basal grade of Poaceae had been included in bamboos at one
time or another. Clark and Triplett (2006) discussed relationships within the
subfamily, previously divided into the woody Bambuseae and the herbaceous
Olyreae. However, the woody temperate bamboo group may be sister to the rest of
the family; the monotypic Buergersiochloa, from New Guinea, is a member
of the monophyletic and otherwise entirely New World woody bamboo group, and the
Olyreae are derived (e.g. Bouchenak-Khelladi 2008). Sungkaew et al. (2009; five
plastid genes) retreived the relationships [Arundinarieae [Olyreae [Neotropical
(strictly) Bambuseae + Paleotropical & Austral Bambuseae]]] - and map the
distributions of each of these groups. For a phylogeny of the woody bamboos, but
with rather little resolution, see Clark et al. (2008), of neotropical woody
bamboos, see Clark et al. (2008) and Fisher et al. (2009), of palaeotropical
woody bamboos, see Yang et al. (2008b: resolution o.k., baccate fruit arose in
parallel), of Bambusa and its relatives, see Yang et al. (2010) and Goh
et al. (2010), of dendrocalamua, see Sungkaew et al. (2010), of temperate
bamboos, see Peng et al. (2008), of Arundinarieae, see Zang et al. (2010: again
rather little resolution despite ca 9,000 bp sequences), and of Bambuseae -
Arthrostylidiinae, see Tyrell et al. (2009).
Within Ehrhartoideae, the relationships of Oryzeae
have been much studied (Guo & Ge 2005; L. Tang et al. 2010 and references);
for diversification within Oryza, see Zou et al. (2008). The first
seedling leaf of Oryzeae does not have a lamina.
For the ndhF gene, structural features of
chloroplast and nuclear genomes, etc.,and the phylogeny of Poöideae, see Davis
and Soreng (2008). It is not certain the the duplication of the ß-amylase gene
is an apomorphy of (many) Poöideae. One of the copies of the gene breaks down
starch into fermentable sugars in the endosperm, while the other is more broadly
expressed in the plant, as it is in other Poaceae (Mason-Gamer 2005). For the
expansion of the inverted repeat in Poöideae at the SSC/IRa boundary, see Saski
et al. (2007). Soreng and Davis (2000) outlined relationships in Poöideae.
Within Poöideae, a number of taxa show complex reticulating patterns of
relationships; for those in Triticeae in particular, see G. Petersen et al.
(2006a) and Mason-Gamer (2008) and references. For relationships and morphology
in Phaenospermateae (inc. Duthieae), see Schneider et al. (2011);
Phaenosperma itself is a very distinct grass previously included in
Bambusoideae. For a phylogeny of Poeae, which should now include Aveneae, see
Quintinar et al. (2007, also Döring et al. 2007; Soreng et al. 2007; Saarela et
al. 2010), for that of Poa, see Gillespie and Soreng (2005), Gillespie et
al. (2009) and Soreng et al. (2010). See also Gillespie et al. (2008, 2010) for
relationships in Poinae, Quintanar et al. (2010) for Koeleriinae, Essi et al.
(2008) for relationships around Briza, and Consaul et al. (2010) for
polyploid speciation in Puccinellia. For a phylogeny of Stipeae in which
Macrochloa may be sister to the rest of the tribe and there are later
parallel diversifications in the Old and New Worlds - characters traditonally
thought to be phylogenetically importamnt appear not to be so - see Romaschenko
et al. (2007, esp. 2008, 2009, 2010; Jacobs et al. 2008; Barkworth et al. 2008),
for New World Stipeae, see Ciadella et al. (2010: but sampling). Winterfeld
(2006) discussed cytogenetic evolution, mainly in the old Aveneae. Inda et al.
(2008a) discuss the biogeography of Loliinae, which seems to have involved
multiple dispersal events from a centre in the Mediterranean region over the
last ca 13 million years. There are several papers on Poooideae in Aliso
23: 335-471. 2008. which should also be consulted, and see Schneider et al.
(2009) for relationships within the whole subfamily.
Classification. For the basic classification
of the family, see the Grass Phylogeny Working Group (2001), although there has
been considerable recent change in detail. See also the World
Checklist of Monocots for a checklist of grasses; Grassworld,
moderated by B. K. Simon, has just started up; Watson and Dallwitz (1992b
onwards) includes generic treatments, etc.
Peterson et al. (2010) provide a detailed
suprageneric classification of Chloridoideae (see also Columbus et al. 2010 for
Muhlenbergia), while Sánchez-Ken and Clark (2010) outline a tribal
classification for a Panicoideae in the broad sense that now include
Centothecoideae. Setaria (Panicoideae) will have to be dismembered
(Kellogg et al. 2009), Panicum itself is getting pulverized, perhaps
overly much so, (e.g. Sede et al. 2008, 2009b; Zuloaga et al. 2010);
Panicum s.l. has about 500 species, s. str. ca 100 species, while
Dicanthelium has about 55 species - see e.g. Zuloaga et al. (2007).
Cenchrus is to include Pennisetum (Chemisquy et al. 2010) and
Muhlenbergia is also to be slightly expanded (Peterson et al. 2010b).
Linder et al. (2010) offer a subfamilial classification of Danthonioideae;
generic limits are difficult there and there has been some confusing
hybridization (Pirie et al. 2009; Humphreys et al. 2010a).
For generic delimitation in the temperate bamboos,
see Peng et al. (2008), in the palaeotropical woody bamboos, see Yang et al.
(2008b) and in Bambusa and its relatives, see Yang et al. (2010). There
are also generic problems in Bambusoideae-Arundinarieae (Zeng et al. 2010) and
-Bambuseae-Arthrostylidiinae (Tyrell et al. 2009); Chusquea must include
Neurolepis (Fisher et al. 2009). Schneider et al. (2009) ouline tribal
limits within Poöideae. For a catalogue of New World Poöideae, see Soreng et al.
(2003). Hybridisation, introgression, and polyploidy are rife in
Poöideae-Triticeae (e.g. G. Petersen et al. 2006a; Mason-Gamer 2008), which
include a number of important grain genera such as Triticum,
Hordeum, etc. Genera are certainly not monophyletic here, but are based
on different genome combinations that are (hopefully) correlated with
morphological variation (Dewey 1984; Löve 1984); Barkworth (2000) summmarises
the history of the classification of this group.
Apparently the earliest name for Chloridoideae is
Chondrosoideae Link, which is a sort of resurrection name - Googling it (as of
3.vii.2007) returned only Thorne and Reveal (2007), apparently the only people
to have used it for some time, and about 42,100 returns for Chloridoideae. Two
cheers for priority!
Botanical Trivia. Woody bamboos, for example
Chusquea, may have a hundred or so branches at a node, produced by a
combination of multiple buds and axillary shoots with very short internodes, all
nodes producing branches (see e.g. Judziewicz et al. 1999).
CLASSIFICATION (APG)
POACEAE Barnhart, nom. cons.//GRAMINEAE
Jussieu, nom. cons. et nom. alt.
(Aerial branching + [?level]); vesicular-arbuscular mycorrhizae +; 3 desoxyanthocyanins,
flavone 5- and C-glycosides, tricin, flavonoid sulphates, (cyanogenic glycosides) +; primary cell wall rich in arabinoxylans, pectin 10³%, xyloglucans lacking fucose; lignins acylated with p-coumarates
[?level]; sieve tube plastids
also with rod-shaped protein bodies, P-proteins 0; arm and fusoid cells
+; cuticle waxes as aggregated rodlets; stomatal subsidiary cellsconical
to dome-shaped; microhairs bicellular; leaves pseudopetiolate,
supervolute(-plicate), midrib +; two adaxial outer T distinct, abaxial
smaller; A centrifixed [?level]; pollen grains central in loculus, with
operculum, wall without
scrobiculi, with intraexinous channels; G (open in development), style
solid [?level]; ovule single, central, amphitropous or hemianatropous,
crassinucellate, [funicle short], micropyle
endostomal; supernumerary antipodals +; fruit an
achene, the testa closely adherent to pericarp [= caryopsis], hilum long
[reverses]; peripheral layer of endosperm meristematic, embryo lateral,
long, well differentiated,
plumule lateral; primary root 0, collar [epiblast, the ligule of the
cotyledon] conspicuous; n = ?; expansion of the inverted repeat
[level?], chloroplast genome with [third!] trnT inversion in the single-copy region, only 17 introns [that in clpP absent], loss of accD, ycf1, ycf2 genes, duplication of AP1/FUL genes [= FUL1 and FUL2], rpoC2 gene insert, rps14 gene to nucleus, pseudogene remaining in mitochondrion, intragenomic translocation of chloroplast rpl23 gene.

1. Anomochlooideae Potzdal
(Leaves spiral -
Streptochaeta); pseudopetiole with an apical (and basal) pulvinus; ligule
as a fringe of hairs; inflorescence branches cymose, two "bracts" along each
branch unit, two more "bracts" below each flower; flowers perfact; P 2 (3) + 3;
or flowers spirally arrranged along racemose axis, with several spiral "bracts"
below each flower, = T, possibly 3 + 3, the latter coriaceous; A (4 -
Anomochloa), sub-basifixed, basally connate, not dangling, [anthers
latrorse, wall development of the Reduced type, endothecium lacking thickenings;
microsporogenesis simultaneous; stigma not plumose - all Streptochaeta];
21bp [long] subrepeats in rpoC2 gene insert; n = 11, 18; first seedling
leaf lacking lamina.
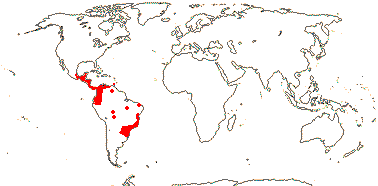
2/4. Central America to S.E. Brasil, scattered, forests (map: from Judziewicz
et al. 1999).
Synonymy: Anomochloaceae Nakai, Streptochaetaceae Nakai
[Pharoideae [Puelioideae [PACMAD + BEP clades]]] / the spikelet clade:
leaves with ± membranous ligules, whether or not also ciliate; inflorescence of
laterally compressed, racemose, pedunculate spikelets, with two basal glumes [sterile bracts = spikelet bract + prophyll],
flowers few, two-ranked, each with lemma and palea [?= bract and 2
adaxial connate outer-whorl tepals], inverted, lodicules [= inner whorl/C?] 3 [median member adaxial];
n = 12; 1 bp deletion in the 3' end of the mat K gene, loss of
rpoC1 gene, 39bp subrepeats in rpoC2 gene insert.
Evolution. Bouchenak-Khelladi et al. (2009,
see also 2010a) estimated that the spikelet clade originated ca 75 million years
ago in the Late Cretaceous, (83-)67(-55) million years, while Bouchenak-Khelladi
et al. (2010c) give an estimate of (83-)67(-55) million years.
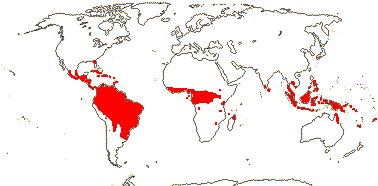
2. Pharoideae L. G. Clark & Judziewicz
Microhairs
0; leaves resupinate, lateral veins oblique; plants
monoecious; spikelets 1-flowered; staminate flowers: A 6, anthers
latrorse, wall of the Reduced type, endothecium lacking thickenings [both
Pharus]; carpellate flowers: style solid; micropyle bistomal
[Pharus]; coleoptile [= first seedling leaf] with lamina.
3/14. Pantropical, in forests (map: from Judziewicz 1987; Judziewicz et al.
1999).
Synonymy: Pharaceae Herter
[Puelioideae / the bistigmatic clade: phytoliths
saddle-shaped; spikelets disarticulating above the glumes; anthers
versatile[?]; pollen grains peripheral in loculus; stigmas 2, two
orders of stigmatic branching; 15bp ndhF insertion.
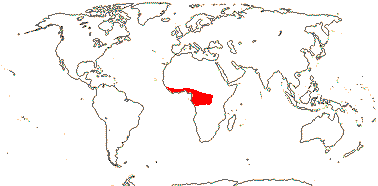
3. Puelioideae L. G. Clark, M. Kobay., S.
Mathews, Spangler & E. A. Kellogg
Characters?; flowers
perfect; A 6; seedling leaf unknown.
2/11. Tropical Africa (map: from Emmet Judziewicz, pers. comm.).
[PACMAD + BEP clades]: (benzoxazinoids, ergot alkaloids [latter synthesized
by endophytes] +); arm and fusoid cells 0; foliar cross veins 0; pseudopetiole 0 flower type?; C/lodicules 2; A 3; G 2, styles separate; antipodal cells
proliferating; x = 12; genome duplication, 15 bp insertion in ndhF
gene, disease resistance by the Hm 1 gene.
These are mostly non-forest grasses.
[Aristidoideae [Panicoideae ]] / PACMAD clade: ligule often of hairs; phytoliths
dumb-bell-shaped; mesocotyl internode elongated, epiblast 0; extension of
ndhF gene from the short single copy region into the inverted repeat.
Evolution. Bouchenak-Khelladi et al. (2009,
see also 2010a) suggest that the PACMAD clade diversified towards the end of the
Eocene some 45-37 million years ago.
4. Aristidoideae Caro
(C4
photosynthesis); ligule with line of hairs; spikelet elongated-cylindrical,
disarticulating above glume; lemma awn trifid, or 3 (1) awns, with basal column;
callus sharp; n = 11, 12; germination flap +; C4 photosynthesis
prevalent.
3/349: Aristida (250-290), Stipagrostis (50). Warm temperate,
few in Europe.
[Panicoideae ]: (C4 photosynthesis); 6 bp insertion in the 3' end
of the mat K gene [?whole clade]
5. Panicoideae Link (includes Centothecoideae
Soderstrom - see Sánchez-Ken & Clark 2010)
(Culms branched);
(fusoid cells +); microhairs elongated, with slender, thin-walled cap cells
["panicoid type"]; (mesophyll differentiated into palisade and spongy tissues;
chlorenchyma cells lobed [cf. arm cells]); culms usually solid; spikelet
development basipetal, dorsally compressed, rachilla 0, 2-flowered, lower flower
staminate or sterile [gynoecial cell death caused by Tasselseed2],
spikelet dispersed as a 1-seeded unit by disarticulation below the glumes;
(style +); hilum non-linear; overlapping embryonic leaf margins; C4
photosynthesis common; starch grains simple; 5 bp insertion in the rpl16
intron; n = (5, 7) 9 [Paniceae], 10 (11, 12, 14); (epiblast +), germination flap
+; rps14 pseudogene lost.
218/3236: Panicum (500 s.l., but polyphyletic, 100 s. str.,
Dicanthelium [55] - see e.g. Zuloaga et al. 2007), Paspalum (330),
Cenchrus (105: inc. Pennisetum), Andropogon (100),
Panicum s. str. (100), Dicanthelium (55), Eriachne (40).
Tropics to temperate.
Synonymy: Andropogonaceae Martinov, Arundinellaceae Herter,
Cenchraceae Link, Panicaceae Berchtold & J. Presl,
Paspalaceae Link, Saccharaceae Berchtold & J. Presl,
Zeaceae A. Kerner
: ?
[Arundinoideae + Micrairoideae]: ?
6. Arundinoideae Burmeister
Microhairs elongated,
with slender, thin-walled cap cells ["panicoid type"]; hilum short; n = 6, 9,
12.
14/45. Temperate to tropical, hydrophytic to xerophytic.
The exact contents of this subfamily are still
unclear.
Synonymy: Arundinaceae Döll
7. Micrairoideae Pilger
Stomata with dome-shaped
subsidiary cells; ligule with fringe of hairs; lemma awn +/0; embryo small,
starch grains simple; n = 10; germination flap +; (C4 photosynthesis
- Eriachneae).
8/188: Isachne (100), Eriachne (35). Tropics.
Micraira has spirally arranged leaves and at
least some species are resurrection plants.
For further information, see Sánchez-Ken et al.
(2007).
[Danthonioideae + Chloridoideae]: hilum short, punctate.
8. Danthonioideae Barker & Linder
Prophylls bilobed
[?distribution]; leaf blades symmetrical, (not Merxmuellera); lemma awn
trifid, or 3 awns; bases of styles well apart; synergid cells haustorial; n = 6,
7, 9.
17/281: Danthonia (100), Rytidosperma (90). Widespread, esp.
Southern Hemisphere, few Southeast Asia-Malesian.
9. Chloridoideae Beilschmied
C4 PCK
subtype (phosphoenolpyruvate carboxykinase) + (0); microhairs usu. with ±
hemispherical and thick-walled distal cells and long base cell, latter with
internal membranes and secretory ["chloridoid type"]; leaf blades symmetrical
(Merxmullera - not); spikelets disarticulating above the glumes; embryo
with an epiblast, mesocotyl +, leaf margins not overlapping; 4 bp insertion in
the rpl16 intron; n = (6-8) 9, 10; C4 photosynthesis
prevalent.
130/1607: Eragrostis (300), Muhlenbergia (155),
Sporobolus (160), Chloris (55). Tropical to warm temperate, more
or less dry environments especially in Africa and Australia.
Synonymy: Chloridaceae Berchtold & J. Presl, Cynodontaceae
Link, Eragrostidaceae Herter, Lepturaceae Herter,
Pappophoraceae Herter, Spartinaceae Link, Sporobolaceae
Herter, nom. inval., Zoysiaceae Link
[Panicoideae + Centothecoideae] Arundinoideae + Chloridoideae: 6 bp
insertion in the 3' end of the mat K gene.
Arundinoideae + Chloridoideae + Aristidoideae + Danthonioideae: ligule
hairy; lemma awned; starch grains compound.
Panicoideae + Centothecoideae: hilum non-linear; overlapping embryonic
leaf margins.
The first character could also be used to unite
Panicoideae + Arundinoideae + Centothecoideae + Chloridoideae.
[Ehrhartoideae [Bambusoideae + Poöideae]] / BEP clade: endosperm softness
gene +, [?embryo short]; x = 12.
10. Ehrhartoideae Link
(Arm cells + - Oryzeae);
(longitudinal walls of epidermal cell straight); (microhairs 0); spikelet
deveklopmental basipetal, with only one carpellate floret fertile and with basal
carpellate or sterile florets, glumes very small; A (1-)6, styles separate
almost from the very base; n = (10, 15); (roots at scutellar node -
Ehrharta).
17/111: Oryza (20), Leersia (20). Widespread, esp. S.
hemisphere.
Synonymy: Ehrhartaceae Link, Oryzaceae Berchtold & J.
Presl
[Bambusoideae + Poöideae]: embryo morphology [that of Brachyelytrum is
like Bambusoideae...], embryonic leaf margins overlapping.
Evolution. Divergence &
Distribution. Wu et al. (2012) suggest that this clade diverged
(48.8-)42.8(-36.6) million years.
11. Bambusoideae Luersson
Woody; culms often
branched; fusoid cells and strongly asymmetrically invaginated arm and fusioid
cells +; microhairs elongated, with slender, thin-walled cap cells ["panicoid
type"]; (multiple buds per node); leaves pseudopetiolate, often with inner and
also outer ligules, culm leaves often very different from the others; (lodicules
3); A (2-)6(-140), (basally connate); stigmas (1-)2-3; (fruit a berry); first
seedling leaf without lamina; n = 7, 9-12, much polyploidy.
84-101/1470. Bambusa (120), Chusquea (200), Sasa (60),
Phyllostachys (55), Arundinaria (50). Tropical to temperate, often
in forests (map: see Judziewicz et al. 1999; Sungkaew et al. 2009).
Synonymy: Bambusaceae Berchtold & J. Presl, Olyraceae
Berchtold & J. Presl, Parianaceae Nakai
12. Poöideae Bentham
Epichloe endophytes pervasive; aerial branching at most rare;
fructose oligosaccharides in stem; root epidermal cell division forming
trichoblast/atrichoblast pair asymmetric; longitudinal walls of epidermal cells
straight [?level]; lemma usually with 5 nerves; lodicules at most slightly
vascularized; styles separate almost from the very base; (postament +); hilum
often short; (endosperm with some non-starch soluble storage polysaccharides);
embryo small, epiblast +, scutellar cleft 0 [scutellum not peltate], mesocotyl
0; n = (2, 4-13); duplication of the ß-amylase gene.
179/3850. Festuca (470: inc. Lolium), Poa (200),
Stipa (300), Calamagrostis (230), Agrostis (220),
Elymus (150), Bromus (100). Largely North Temperate.
1. Brachyelytreae Ohwi
Stomata subsidiary cells
with parallel sides; n = 11.
1/3. Eastern Asia, E. North America.
[Nardeae [Phaenospermateae + The Rest]]: primary inflorescence branches
2-ranked; embryo lacking scutellar cleft, embryonic leaf margins
non-overlapping.
2. Nardeae Koch
Lodicules 0; style and
stigma 1; n = 10, 13.
2/2. Europe.
Synonymy: Nardaceae Martynov
[Phaenospermateae + The Rest]: microhairs 0 (+ - some Stipeae); (stomata
subsidiary cells with parallel sides); n = 7 [chromosomes "large"].
3. Phaenospermateae
21 bp insertion in in
rpl32-trnL; n = 7, 12.
7/11. Central to East Asia, also Australia, Mexico, Balkans, Caucasus;
scatttered.
Synonymy: Aegilopaceae Martynov, Agrostidaceae Berchtold &
J. Presl, Alopecuraceae Martynov, Anthoxanthaceae Link,
Avenaceae Martynov, Bromaceae Berchtold & J. Presl,
Chaeturaceae Link, Cynosuraceae Link, Festucaceae Sprengel,
Glyceriaceae link, Holcaceae Link, Hordeaceae Berchtold
& J. Presl, Laguraceae Link, Loliaceae Link, Melicaceae
Martynov, Miliaceae Link, Phalaridaceae link, Phleaceae
Link, Sesleriaceae Döll, Stipaceae Berchtold & J. Presl,
Triticaceae Link
References
Extracted from Angiosperm Phylogeny references compiled by Peter Stevens
Aliscioni, S. S. [et al.
2003], Giussani, L. M., Zuloaga, F. O., & Kellogg, E. A. 2003. A
molecular phylogeny of Panicum (Poaceae: Paniceae): Tests of monophyly
and phylogenetic placement within the Panicoideae. American J. Bot. 90:
796-821.
Amarasinghe, V., & Watson, L. 1988. Comparative
ultrastructure of microhairs in grasses. Bot J. Linnean Society 98:
303-319.
Amarasinghe, V., & Watson, L. 1990. Taxonomic
significance of microhair morphology in the genus Eragrostis Beauv.
(Poaceae). Taxon 39: 59-65.
Arakaki, M. [et al. 2011], Christin, P.-A.,
Nyffeler, R., Lendel, A., Eggli, U., Ogburn, R. M., Spriggs, E., Moore, M. J., &
Edwards, E. J. 2011. Contemporaneous and recent radiations of the world's major
suculent lineages. Proc. National Acad. Sci. U.S.A. 108: 8379-8384.
Barker, N. P. [et al. 2007a], Galley, C., Verboom,
G. A., Mafa, P., Gilbert, M., & Linder, H. P. 2007a. The phylogeny of the
austral grass subfamily Danthonioideae: Evidence from multiple data sets. Plant
Syst. Evol. 264: 135-156.
Barkworth, M. E. 2000. Changing perceptions of the
Triticeae. Pp. 110-120, in Jacobs, S. W. L., & Everett, J. (eds), Grasses:
Systematics and Evolution. CSIRO, Melbourne.
Barkworth, M. E. [et al. 2008], Arriaga, M. O.,
Smith, J. F., Jacobs, S. W. L., Valdés-Reyna, J., & Bushman, B. S. 2008.
Molecules and morphology in South American Stipeae (Poaceae). Syst. Bot.
33: 719-731.
Bertin, C. [et al. 2007], Weston, L. A., Huang, T.,
Jander, G., Owens, T., Meinwald, J., & Schroeder, F. C. 2007. Grass roots
chemistry: meta-Tyrosine, an herbicidal nonprotein amino acid. Proc.
National Acad. Sci. U.S.A. 104: 16964-16969.
Blanc, G., & Wolfe, K. H. 2004. Widespread
paleopolyploidy in model plant species inferred from age distributions of
duplicate genes. Plant Cell 16: 1667-1678.
Bläsing, O. E. [et al. 2000], Westhoff, P., &
Svensson, P. 2000. Evolution of C4 phosphoenolpyruvate
carboxylase in Flaveria, a conserved serine residue in the
carboxy-terminal part of the enzyme is a major determinant for C4-specific
characteristics. J. Biol. Chem. 275: 27917-27923.
Bocherens, H. [et al. 1993], Friis, E. M.,
Mariotti, A., & Pedersen, K. R. 1993. Carbon isotope abundance in Mesozoic
and Cenozoic fossil plants: Palaeoecological implications. Lethaia 26:
347-358.
Bond, W. J. 2008. What limits trees in C4
grasslands and savannas? Ann. Review Ecol. Syst. 39: 641-659.
Bouchenak-Khelladi, Y. [et al. 2008], Salamin, N.,
Savolainen, V., Forest, F., van der Bank, M.,
Chase, M. W., & Hodkinson, T. R. 2008. Large multi-gene phylogenetic trees
of the grasses (Poaceae): Progress towards complete tribal and generic level
sampling. Mol. Phyl. Evol. 47: 488-505.
Bouchenak-Khelladi, Y. [et al.
2009], Verboom, G. A., Hodkinson, T. R., Salamin, N., Francois, O., Chongaile,
G. N., & Savolainen, V. 2009. The origins and diversification
of C4 grasses and savanna-adapted ungulates. Global Change Biol.
15: 2397-2417.
Bouchenak-Khelladi, Y. [et al. 2010a], Verboom, G.
A., Savolainen, V., & Hodkinson, T. R. 2010a. Biogeography of the grasses
(Poaceae): A phylogenetic approach to reveal evolutionary history in
geographical space and geological time. Biol. J. Linnean Soc. 162:
543-557.
Bouchenak-Khelladi, Y. [et al.
2010c], Savolainen, V., & Hodkinson, T. R. 2010c. Diversification
of the grasses (Poaceae): A phylogenetic approach to reveal macro-evolutionary
patterns. Pp. 451-475, in Seberg, O., Petersen, G., Barfod, A. S., & Davis,
J. I. (eds), Diversity, Phylogeny, and Evolution in the Monocotyledons. Aarhus University
Press, Århus.
Boykin, L. M. [et al. 2008], Pockman, W. T., &
Lowrey, T. K. 2008. Leaf anatomy of Orcuttieae (Poaceae: Chloridoideae): More
evidence of C4 photosynthesis without Kranz anatomy. Madroño
55: 143-150.
Bremer, K. 2002. Gondwanan evolution of the grass
alliance of families (Poales). Evolution 56: 1374-1387.
Brown, N. J. [et al. 2011], Newell, C. A., Stanley,
S., Chen, J. E., Perrin, A. J., Kajala, K., & Hibberd, J. M. 2011.
Independent and parallel recruitment of preexisting mechanisms underlying C4
photosynthesis. Science 331: 1436-1439.
Cahoon, A. B. [et al. 2010], Sharpe, R. M.,
Mysayphonh, C., Thompson, E. J., Ward, A. D., & Lin, A. 2010. The complete
chloroplast genome of tall fescue (Lolium arundinaceum; Poaceae) and
comparison of whole plastomes from the family Poaceae. American J. Bot.
97: 49-58.
Cerros-Tlatilpa, R., & Columbus, J. T. 2009. C3
photosynthesis in Aristida longifolia: Implication for photosynthetic
diversification in Aristidoideae (Poaceae). American J. Bot. 96:
1379-1387.
Chemisquy, M. A. [et al. 2010], Giussani, L. M.,
Scataglini, M. A., Kellogg, E. A., & Morrone, O. 2010. Phylogenetic studies
favour the unification of Pennisetum, Cenchrus and Odontelytrum
(Poaceae): A combined nuclear, plastid and morphological analysis, and
nomenclatural combinations in Cenchrus. Ann. Bot. 106: 107-130.
Christin, P. A., & Besnard, G. 2009. Two
independent C4 origins in Aristidoideae (Poaceae) revealed by the
recruitment of distinct phosphoenolpyruvate carboxylase genes. American J.
Bot. 96: 2234-2239.
Christin, P.-A. [et al. 2008], Besnard, G.,
Samaritani, E., Duvall, M. R., Hodkinson, T. R., Savolainen, V., & Salamin,
N. 2008. Oligocene CO2 decline promoted C4 photosynthesis
in grasses. Curr. Biol. 18: 37-43.
Christin, P.-A. [et al.
2009a], Petitpierre, B., Salamin, N., Büchi, L., & Besnard, G. 2009a. Evolution
of C4 phosphoenolpyruvate carboxykinase in grasses, from
genotype to phenotype. Mol. Biol.
Evol. 26: 357-365.
Ciadella, A. M. [et al. 2010],
Salariato, D. L., Aagesen, L., Giussani, L. M., Zuloaga, F. O., & Morrone,
O. 2010. Phylogeny of the New World Stipeae (Poaceae): An evaluation of the
monophyly of Aciachne and Amelichloa. Cladistics 26:
563-578.
Clark, L. G. 1997. Bamboos: The centrepiece of the
grass family. Pp. 237-248, in Chapman, G. P. (ed.), The Bamboos.
Academic Press, London.
Clark, L. G., & Triplett, J. K. 2006. Phylogeny
of the Bambusoideae (Poaceae): An update. P. 212, in Botany 2006 - Looking
to the Future - Conserving the Past. [Abstracts: Botanical Society of America, etc.]
Clark, L. G. [et al. 2008], Dransfield, S., Triplett,
J., & Sánchez-Ken, J. G. 2007 [2008]. Phylogenetic relationships amopng the
one-flowered, determinate genera of Bambuseae (Poaceae: Bambusoideae). Pp.
315-332, in Columbus, J. T., Friar, E. A., Porter, J. M., Prince, L. M., &
Simpson, M. G. (eds), Monocots: Comparative Biology and Evolution. Poales..
Rancho Santa
Ana Botanical Garden,
Claremont, Ca.
[Aliso 23: 315-332.]
Columbus, J. T.
[et al. 2010], Peterson, P. M., Refulio-Rodríguez, N. F., Cerros Tlatilpa, R.,
& Kinney, M. S. 2010. phylogenetics of Muhlenbergiinae (Poaceae:
Chloridoideae, Cynodonteae) based on ITS and trnL-F sequences. Pp.
477-495, in Seberg, O., Petersen, G., Barfod, A. S., & Davis, J. I. (eds), Diversity,
Phylogeny, and Evolution in the Monocotyledons. Aarhus University
Press, Århus.
Consaul, L. L. [et al. 2010], Gillespie, L. G.,
& Waterway, m. J. 2010. Polyploid speciation and evolution in Arctic Puccinellia
(Poaceae: Puccinelliinae) - a review. pp. 645-662, in Seberg, O., Petersen, G.,
Barfod, A. S., & Davis, J. I. (eds), Diversity, Phylogeny, and Evolution
in the Monocotyledons. Aarhus
University Press, Århus.
Cornwell, W. K. [et al. 2008], Cornelissen, J. H.
C., Amatangelo, K., Dorrepaal, E., Eviner, V. T., Godoy, O., S. E. Hobbie, S.
E., Hoorens, B., Kurokawa, H., Perez Harguindeguy, N., Quested, H. M.,
Santiago, L. S., Wardle, D. A., Wright, I. J., Aerts, R., Allison, S. D., van
Bodegom, P., Brovkin, V., Chatain, A., Callaghan, T., Díaz, S., Garnier, E.,
Gurvich, D. E., Kazakou, E., Klein, J. A., Read, J., Reich, P. B., Soudzilovskaia,
N. A., Vaieretti, M. V., & Westoby, M. 2008. Plant species traits are the
predominant control on litter decomposition rates within biomes worldwide. Ecol.
Lett. 11: 1065-1071.
Crepet, W. L., & Feldman, G. D. 1991. The
earliest remains of grasses in the fossil record. American J. Bot. 78:
1010-1014.
Crins, W. J. 1991. The genera of Paniceae
(Gramineae: Panicoideae) in the Southeastern United States.
J. Arnold Arbor. Suppl. Ser. 1: 171-312.
Davidse, G. 1987. Fruit dispersal in the Poaceae.
Pp. 143-155, in Soderstrom, T. R., Hilu, K. W., Campbell, C. S., &
Barkworth, M. E. (eds), Grass Systematics and Evolution. Smithsonian
Institution Press, Washington,
D.C.
Davis, J. I., & Soreng, R. J. 2007 [2008]. A
preliminary phylogenetic analysis of the grass subfamily Poöideae (Poaceae),
with attention to structural features of the plastid and nuclear genomes,
including an intron loss in GBSSI. Pp. 335-348, in Columbus, J. T., Friar, E.
A., Porter, J. M., Prince, L. M., & Simpson, M. G. (eds), Monocots:
Comparative Biology and Evolution. Poales.. Rancho Santa Ana
Botanical Garden, Claremont, Ca. [Aliso 23: 335-348.]
Dewey, D. R. 1984. The
genomic system of classification as a guide to intergeneric hybridization with
the perennial Triticeae. Pp. 209-279, in Gustafson, J. P. (ed.), Gene
Manipulation in Plant Imporvement. Plenum, New York.
Dhillon, T. [et al. 2010], Pearce, S. P.,
Stockinger, E. J., Distelfeld, A., Li, C., Vashegyi, A. I., Vágújfalvi, A.,
Galiba, G., & Dubcovsky, J. 2010. Regulation of freezing tolerance and
flowering in temperate cereals: The VRN-1 connection. Plant. Physiol.
153: 1846-1858.
Donadío, S. [et al. 2009], Giussani, L., Kellogg,
E. A., Zuolaga, F.O., & Morrone, O. 2009. A preliminary molecular phylogeny
of Pennisetum and Cenchrus (Poaceae - Paniceae) based on the trnL-F,
rpl16 chloroplast markers. Taxon 58: 392-404.
Döring, E. [et al. 2007], Schneider, J., Hilu, K.
W., & Röser, M. 2007. Phylogenetic relationsnships in the Aveneae/Poeae
complex (Poöideae, Poaceae). Kew
Bull. 62: 407-424.
Doust, A. N., & Kellogg, E. A. 2002.
Integrating phylogeny, developmental morphology and genetics: A case study of
inflorescence evolution in the 'bristle grass' clade (Panicoideae: Poaceae). Pp
298-314, in Cronk, Q. C. B., Bateman, R. M., & Hawkins, J. A. (eds), Developmental
Genetics and Plant Evolution. Taylor and
Francis, London.
Doust, A. N. [et al. 2007], Penly, A. M., Jacobs,
S. W. L., & Kellogg, E. A. 2007. Congruence, conflict, and polyploidization
shown by nuclear and chloroplast markers in the monophyletic "bristle
clade" (Paniceae, Panicoideae, Poaceae). Syst. Bot. 32: 531-544.
Doyle, J. J. [et al. 1992], Davis, J. I., Soreng,
R. J., Garvin, D., & Anderson, M. J. 1992. Chloroplast DNA inversions and
the origin of the grass family (Poaceae). Proc. National Acad. Sci. U.S.A. 89: 7722-7726.
Duvall, M. R. [et al. 2007],
Leseberg, C. H., & Grennan, C. P. 2007. Comparison of whole chloroplast
genomes in grasses (Poaceae); evolutionary insights related to Coix
lacrymajobi, Microcalamus convallarioides, Puelia olyriformis
and Joinvillea plicata. P. 284, in Plant Biology and Botany 2007.
Program and Abstract Book. Chicago.
Duvall, M. R. [et al. 2008a], Davis, J. I., Clark,
L. G., Noll, J. D., Goldman, D. H., & Sánchez-Ken, G. 2007 [2008a].
Phylogeny of the grasses (Poaceae) revisited. Pp. 237-247, in Columbus, J. T.,
Friar, E. A., Porter, J. M., Prince, L. M., & Simpson, M. G. (eds), Monocots:
Comparative Biology and Evolution. Poales.. Rancho Santa Ana
Botanical Garden, Claremont, Ca. [Aliso 23: 237-247.]
Duvall, M. R. [et al. 2010],
Leseberg, C. H., Grennan, C. P., & Morris, l. M. 2010. Molecular
evolution and phylogenetics of complete chloroplast genomes in Poaceae. Pp.
437-450, in Seberg, O., Petersen, G., Barfod, A. S., & Davis, J. I. (eds), Diversity,
Phylogeny, and Evolution in the Monocotyledons. Aarhus University
Press, Århus.
Edwards, E. J. 2009. C4 the straw man? Evolution of
cold tolerance better explains global distribution of C3/C4 grasslands. P. 109,
in Botany and Mycology 2009. Snowbird, Utah July 25-29. Abstract Book.
Edwards, E. J., & Smith, S. A. 2010.
Phylogenetic analyses reveal the shady history of C4 grasses. Proc.
National Acad. Sci. U.S.A. 107: 2532-2537.
Edwards, E. J.[et al. 2007], Still, C. J., &
Donoghue, M. J. 2007. The relevance of phylogeny to studies of global change. Trends
Ecol. Evol. 22: 243-249.
Edwards, E. J. [et al. 2010], Osborne, C. P.,
Strömberg, C. A. E., Smith, S. A., & C4 Grasses Consortium.
2010. The origins of C4 grasslands: Integrating evolutionary and
ecosystem science. Science 328: 587-591.
Ehleringer, J. R. [et al. 1997], Cerling, T. E.,
& Helliker, B. R. 1997. C4photosynthesis, atmospheric CO2,
and climate. Oecologia 112: 285-299.
Essi, L. [et al. 2008], Longhi-Wagner, H. M., &
Teixeira de Souza-Chies, T. 2008. Phylogenetic analysis of the Briza
complex (Poaceae). Mol. Phyl. Evol. 47: 1018-1029.
Fisher, A. [et al. 2009], Triplett, J. K., Ho,
C.-S., Schiller, A. D., Oltrogge, K. A., Schroder, E. S., Kelchner, S. A.,
& Clark, L. G. 2009. Paraphyly in the bamboo subtribe Chusqueinae (Poaceae:
Bambusoideae) and a revised infrageneric classification for Chusquea. Syst.
Bot. 34: 673-683.
Fox, D. L., & Koch, P. L. 2004. Carbon and
oxygen isotopic variability in Neogene paleosol carbonates: Constraints on the
evolution of C4-grasslands of the Great Plains, USA. Palaeogeog.
Palaeoclim. Palaeoecol. 207: 305-329.
Frey, M. [et al. 1997], Chomet, P., Glawischnig,
E., Stettner, C., Grühn, S., Winklmair, A., Eisenreich, W., Bacher, A., Meeley,
R. B., Briggs, S. P., Simcox, K., & Gierl, A. 1997. Analysis of a chemical
plant defense mechanism in grasses. Science 277: 696-699.
Galley, C., & Linder, H.
P. 2007. The phylogeny of the Pentaschistis calde (Danthonioideae,
Poaceae) based on chloroplast DNA, and the evolution and loss of complex
characters. Evolution 61: 864-884.
Gerhart, L. M., & Ward, J. K. 2010. Plant
response to low [CO2] of the past. New Phytol. 188: 674-695.
Gillespie, L. J., & Soreng, R. J. 2005. A
phylogenetic analysis of the bluegrass genus Poa based on cpDNA restriction
site data. Syst. Bot. 30: 84-105.
Gillespie, L. J. [et al. 2008], Soreng, R. J.,
Bull, R. D., Jacobs, S. W. L., & Refulio-Rodruiguez, N. F. 2008.
Phylogenetic relationships in subtribe Poinae (Poaceae, Poeae) based on nuclear
ITS and plastid trnT-trnL-trnF sequences. Botany
86: 938-967.
Gillespie, L. J. [et al.
2009], Soreng, R. J., & Jacobs, S. W. L. 2009. Phylogenetic
relationships of Australian Poa (Poaceae: Poinae), including molecular
evidence for two new genera, Saxipoa and Sylvipoa. Australian
Syst. Bot. 22: 413-436.
Gillespie, L. J. [et al. 2010], Soreng, R. J.,
Paradis, M., & Bull, R. D. 2010. Phylogeny and reticulation in subtribe
Poinae and related subtribes (Poaceae) based on nrITS, ETS, and trnTLF
data. Pp. 589-617, in Seberg, O., Petersen, G., Barfod, A. S., & Davis, J.
I. (eds), Diversity, Phylogeny, and Evolution in the Monocotyledons. Aarhus University
Press, Århus.
Giussani, L. M. [et al. 2001], Cota-Sánchez, J. H.,
Zuloaga, F. O., & Kellogg, E. A. 2001. A molecular phylogeny of the grass
subfamily Panicoideae (Poaceae) shows multiple origins of C4
photosynthesis. American J. Bot. 88: 1993-2012.
Goh, W. L. [et al. 2010], Chandran, S., Lin, R.-S.,
Xia, N.-H., & Wong, K. M. 2010. Phylogenetic relationships among Southeast
Asian climbing bamboos (Poaceae: Bambusoideae) and the Bambusa complex. Mol.
Phyl. Evol. 38: 764-773.
Gómez-Martínez, R., & Culham, A. 2000.
Phylogeny of the subfamily Panicoideae with emphasis on the tribe Paniceae:
Evidence from the trnL-F cpDNA region. Pp. 136-140, in Jacobs, S. W. L.,
& Everett, J. (eds), Grasses: Systematics and Evolution. CSIRO,
Melbourne.
Guisinger, M. M. [et al. 2010], Chumley, T. W.,
Kuehl, J. V., Boore, J. L., & Jansen, R. K. 2010. Implications of the
plastid genome sequence of Typha (Typhaceae, Poales) for understanding
genome evolution in Poaceae. J. Molec. Evol. 70: 149-166.
Guo, Y.-L., & Ge, S. 2005. Molecular phylogeny
of Oryzeae (Poaceae) based on DNA sequences from chloroplast, mitochondrial,
and nuclear genomes. American J. Bot. 92: 1548-1558.
Hall, D. O. [et al. 2000], Scurlock, J. M. O.,
Ojima, D. S., & Parton, W. J. 2000. Grasslands and the global carbon cycle:
Modeling the effects of climate change. Pp. 102-114, in Wigley, T. M. L., &
Schimel, D. S. (eds), The Carbon Cycle. Cambridge
University Press, Cambridge.
Hendry, G. A. F., & Wallace, R. K. 1993. The
origin, distribution and evolutionary significance of fructans. Pp. 119-139, in
Suzuki, M., & Chatterton, J. N. (eds), Science and Technology of
Fructans. CRC Press, Boca Raton,
FLA.
Hilu, K. 2004. Phylogenetics and chromosomal
evolution in the Poaceae (grasses). Australian
J. Bot. 52: 13-22.
Hodkinson, T. R. [et al.
2007], Savolainen, V., Jacobs, S. W. L., Bouchenak-Khelladi, Y., Kinney, M. S.,
& Salamin, N. 2007. Supersizing: Progress in documenting and
understanding grass species richness. Pp. 275-295, in Hodkinson, T. R., &
Parnell. J. A. N. (eds), Reconstructing the Tree of Life: Taxonomy and
Systematics of Species Rich Taxa, CRC Press, Boca Raton, FLA.
[Systematics Association Special Volume Series
72.].
Hodkinson, T. R. [et al.
2008], Salamin, N., Chase, M.W., Bouchenak-Khelladi, Y., Renvoize, S. A., &
Savolainen, V. 2007 [2008]. Large trees, supertreess, and
diversification of the grass family. Pp. 248-258, in Columbus, J. T., Friar, E.
A., Porter, J. M., Prince, L. M., & Simpson, M. G. (eds), Monocots:
Comparative Biology and Evolution. Poales.. Rancho Santa Ana
Botanical Garden, Claremont, Ca. [Aliso 23: 248-258.]
Hultén, E. 1962. The
circumpolar plants. I. Vascular cryptogams. conifers, monocotyledons. Kungl.
Svenska Vetenskap. Handl. ser. 3, 8(5): 1-275.
Humphreys, A. M. [et al. 2010a], Pirie, M. D.,
& Linder, H. P. 2010. A plastid tree can bring order to the chaotic generic
taxonomy of Rytidosperma Steud. s.l. (Poaceae). Molec. Phyl. Evol.
55: 911-928.
Ibrahim, D. G. [et al. 2009], Burke, T., Ripley, B.
S., & Osborne, C. P. 2009. A molecular phylogeny of the genus Allopteropsis
(Panicoideae, Poaceae), suggests an evolutionary reversion from C4
to C3 photosynthesis. Ann. Bot. 103: 127-136.
Inda, L. A. [et al. 2008a], Segarra-Moragues, J.
G., Müller, J., Peterson, P. M., & Catalán, P. 2008a. Dated historical
biogeography of the temperate Loliinae (Poaceae, Pooideae) grasses in the
northern and southern hemispheres. Mol. Phyl. Evol. 46: 932-957.
Ingram, A. L. 2010. Evolution of leaf blade anatomy
in Eragrostis (Poaceae). Syst. Bot. 35: 755-765.
Ingram, A. L. [et al. 2011], Christin, P.-A., &
Osborne, C. P. 2011. Molecular phylogenies disprove a hypothesized C4
reversion in Eragrostis walteri (Poaceae). Ann. Bot. 107:
321-325.
Jacobs, B. F. [et al. 1989], Kingston, J. D., & Jacobs, B. L. 1999.
The origin of grass-dominated ecosystems. Ann. Missouri Bot. Gard. 86: 590-643.
Jacobs, S. W. L. [et al. 2008], Bayer, R., Everett,
J., Arriaga, M. O., Barkworth, M. E., Sabin-Badereau, A., Torres, M. A.,
Vásquez, F. M., & Bagnall, N. 2008. Systematics of the tribe Stipeae using
molecular data. Pp. 49-361, in Columbus, J. T., Friar, E. A., Porter, J. M.,
Prince, L. M., & Simpson, M. G. (eds), Monocots: Comparative Biology and
Evolution. Poales. [Aliso 23: 349-361.]
Jakob, S. S. [et al. 2004], Meister, A., &
Blattner, F. R. 2004. The considerable genome size variation of Hordeum
species (Poaceae) is linked to phylogeny, life form, ecology, and speciation
rates. Mol. Biol. Evol. 21: 860-869.
Janssen, T., & Bremer, K. 2004. The age of
major monocot groups inferred from 800+ rbcL sequences. Bot. J.
Linnean Soc. 146: 385-398.
Judziewicz, E. J. 1987. Taxonomy and Morphology
of the Tribe Phareae (Poaceae: Bambusoideae). Ph. D. Thesis, University of
Wisconsin-Madison.
Judziewicz, E. J., & Clark, L. G. 2007 [2008].
Classification and biogeograpjhy of New World
grasses: Anomochlooideae, Pharoideae, Ehrhartioideae, and Bambusoideae. Pp.
303-314, in Columbus, J. T., Friar, E. A., Porter, J. M., Prince, L. M., &
Simpson, M. G. (eds), Monocots: Comparative Biology and Evolution. Poales..
Rancho Santa Ana Botanical Garden, Claremont, Ca. [Aliso 23: 303-314.]
Judziewicz, E. J., &
Soderstrom, T. R. 1989. Morphological, anatomical and taxonomic studies in Anomochloa
and Streptochaeta (Poaceae: Bambusoideae). Smithsonian Contrib. Bot.
68: 1-52.
Judziewicz, E. J. [et al. 1999], Clark, L. G.,
Londoño, X., & Stern, M. J. 1999. American Bamboos. Smithsonian
Institution, Washington.
Kellogg, E. A. 1999. Phylogenetic aspects of the
evolution of C4 photosynthesis. Pp. 411-444, in Sage, R. F., &
Monson, R. K. (eds), C4 plant biology. Academic Press, San Diego.
Kellogg, E. A. 2000a. The grasses: A case study in
macroevolution. Ann. Rev. Ecol. Syst. 31: 217-38.
Kellogg, E. A. 2000c. Molecular and morphological
evolution in the Andropogoneae. Pp. 149-158, in Jacobs, S. W. L., &
Everett, J. (eds), Grasses: Systematics and Evolution. CSIRO, Melbourne.
Kellogg, E. A. 2001. Evolutionary history of the
grasses. Plant Physiol. 125: 1198-1205.
Kellogg, E. A. [et al. 2009], Aliscioni, S.,
Morrone, O., Pensiero, J., & Zuloaga, F. 2009. A phylogeny of Setaria
(Poaceae, Panicoideae, Paniceae) and related genera based on the chloroplast
gene ndhF. Internat. J. Plant Sci. 170: 117-131.
Kim, C. [et al. 2009], Tang, H., & Paterson, A.
H. 2009. Duplication and divergence of grass genomes: Integrating the
chloridoids. Trop. Plant Biol. 2: 51-62.
Kirpes, C. C. [et al. 1996], Clark, L. G., &
Lersten, N. R. 1996. Systematic significance of pollen arrangement in
microsporangia of Poaceae and Cyperaceae: Review and observations on
representative taxa. American J. Bot. 83: 1609-1622.
Le Roux, L. G., & Kellogg, E. A. 1999. Floral
development and the formation of unisexual spikelets in the Andropogoneae
(Poaceae). American J. Bot. 86: 354-366.
Linder, H. P., & Rudall, P. J. 2005.
Evolutionary history of Poales. Ann. Rev. Ecol. Syst. 36: 107-124.
Linder, H. P. [et al. 2010], Baeza, M., Barker, N.
P., Galley, C., Humphreys, A. M., Lloyd, K. M., Orlovich, D,. A., Pirie, M. D.,
Simon, B. K., Walsh, N., & Verboom, G. A. 2010. A generic classification of
the Danthonioideae (Poaceae). Ann.
Missouri Bot. Gard. 97:
306-364.
Liu, Q. [et al. 2005], Zhao,
N.-X., & Hao, G. 2005. The phylogeny of the Chloridoideae
(Gramineae): A cladistic analysis. J.
Trop. Subtrop. Bot. 13: 432-442.
Liu, Q. [et al. 2010], Zhang,
D. X., & Peterson, P. M. 2010. Lemma micromorphological
characters in the Chloridoideae (Poaceae) optimized on a molecular phylogeny. South
African J. Bot. 76: 196-209.
Livingston, D. P. III [et al. 2009], Hincha, D. K.,
& Heyer, A. G. 2009. Fructan and its relationship to abiotic stress
tolerance in plants. Cell. Molec. Life Sci. 66: 2007-2023.
Lloyd, J., & Farquhar, G. D. 1994. 13C
discrimination during CO2 assimilation by the terrestrial biosphere.
Oecologia 99: 201-215.
Löve, Á, 1984. Conspectus of the Triticeae. Feddes
Repert. 95: 425-451.
Luo, S. [et al. 2011], Peng, J., Li, K., Wang, M.,
& Kuang, H. 2011. Contrasting evolutionary patterns of the Rp1
resistance gene family in different species of Poaceae. Mol. Biol. Evol.
28: 313-325.
McInerney, F. A. [et al. 2011], Strömberg, C. A.
E., & White, J. W. C. 2011. The Neogene transition from C3 to C4
grasslands in North America: Stable carbon
isotope ratios of fossil phytoliths. Paleobiol. 37: 23-49.
Magallón, S. A., & Sanderson, M. J. 2001.
Absolute diversification rates in angiosperm clades. Evolution 55:
1762-1780.
Marchant, A. D., & Briggs, B. G. 2007.
Ecdeiocoleaceae and Joinvilleaceae, sisters to Poaceae (Poales): Evidence from rbcL
and matK data. Telopea 11: 437-450.
Mason-Gamer, R. J. 2005. The ß-amylase genes of
grasses and a phylogenetic analysis of the Triticeae (Poaceae). American J.
Bot. 92: 1045-1058.
Mason-Gamer, R. J. 2008. Allohexaploidy,
introgression, and the complex phylogenetic history of Elymus repens Mol.
Phyl. Evol. 47: 598-611.
Mathews, S., & Sharrock, R. A. 1996. The
phytochrome gene family in grasses (Poaceae): A phylogeny and evidence that grasses
have a subset of the loci found in dicot angiosperms. Mol. Biol. Evol.
13: 1141-1150.
Mathews, S. [et al. 2002], Spangler, R. E.,
Mason-Gamer, R. J., & Kellogg, E. A. 2002. Phylogeny of Andropogoneae
inferred from phytochrome B, GBSSI, and ndhF. Internat. J. Plant Sci. 163:
441-450.
Michelangeli, F. A. [et al.
2002], Davis, J. I., & Stevenson, D. W. 2002. Phylogenetic
relationships among Poaceae and related families as inferred from morphology,
chloroplast structure, and sequence data from the mitochondrial and plastid
genomes. P. 139, in Botany 2002: Botany in the Curriculum, Abstracts. [Madison, Wisconsin.]
Michelangeli, F. A. [et al.
2003], Davis, J. I., & Stevenson, D. W. 2003. Phylogenetic
relationships among Poaceae and related families as inferred from morphology,
inversions in the plastid genome, and sequence data from the mitochondrial and
plastid genomes. American J. Bot. 90: 93-106.
Morris, L. M., & Duval, M. R. 2010. The
chloroplast genome of Anomochloa marantoidea (Anomochlooideae; Poaceae)
comprises a mixture of grass-like and unique features. American J. Bot.
97: 620-627.
Morrone, O. [et al. 2010], Aagesen, L., Scataglini,
M. A., Salariato, D., Denham, S. S., Chemisquy, M. A., Sede, S., Giussani, L.
M., Kellogg, E. A., & Zuloaga, F. O. 2010. Phylogeny of the Paniceae
(Poaceae: Panicoideae) integrating chloroplast DNA sequences and morphology. Cladistics 26: 219.
Ng'uni, D. [et al. 2010],
Geleta, M., Fatih, M., & Bryngelsson, T. 2010. Phylogenetic
analysis of Sorghum based on combined sequence data from cpDNA regions
and ITS generate well-supported trees with two major lineages. Ann. Bot.
105: 471-480.
Osborne, C. P. 2008. Atmosphere, ecology and
evolution: What drove the Miocene expansion of C4 grasslands? J.
Ecol. 96: 35-45.
Osborne, C. P., & Beerling, D. J. 2006.
Nature's green revolution: The remarkable evolutionary rise of C4
plants. Phil. Trans. Roy. Soc. London B, 361: 173-194.
Osborne, C. P., & Freckleton, R. P. 2009.
Ecological selection pressures for C4 photosynthesis in the grasses.
Proc. Roy. Soc. B, 276: 1753-1760.
Pagani, M. [et al. 2009], Caldeira, K., Berner, R.,
& Beerling, D. J. 2009. The role of terrestrial plants in limiting
atmospheric CO2 decline over the past 24 million years. Nature
460: 85-88.
Paterson, A. [et al. 2004], Bowers, J. E., &
Chapman, B. A. 2004. Ancient polyploidization predating divergence of the
cereals, and its consequences for comparative genomics. Proc. National Acad. Sci. U.S.A. 101: 9903-9908.
Peng, S. [et al. 2008], Yang,
H.-Q., & Li, D.-Z. 2008. Highly heterogeneous generic delimitation
within the temperate bamboo clade (Poaceae: Bambusoideae): Evidence from GBSS1
and ITS sequences. Taxon 57: 799-810.
Petersen, G. [et al. 2006a], Seberg, O., Yde, M.,
& Berthelsen, K. 2006a. Phylogenetic relationships of Triticum and Aegilops
and evidence for the origin of the A, B, and D genomes of
common wheat (Triticum aestivum). Mol. Phyl. Evol. 39: 70-82.
Peterson, P. M. [et al. 2009], Romaschenko, K.,
& Johnson, G. 2009. A phylogeny of the Chloridoideae (Poaceae) based on
plastid and nuclear DNA sequence data. P. 194, in Botany and Mycology 2009.
Snowbird, Utah
July 25-29. Abstract Book.
Peterson, P. M. [et al. 2009], Romaschenko, K.,
& Johnson, G. 2009. A phylogeny of the Chloridoideae (Poaceae) based on
plastid and nuclear DNA sequence data. P. 194, in Botany and Mycology 2009.
Snowbird, Utah
July 25-29. Abstract Book.
Peterson, P. M. [et al. 2010a], Romaschenko, K.,
& Johnson, G. 2010a. A classification of the Chloridoideae (Poaceae) based
on multi-gene phylogenetic trees. Mol. Phyl. Evol. 55: 580-598.
Peterson, P. M. [et al. 2010b], Romaschenko, K.,
& Johnson, G. 2010b. A phlogeny and classification of the Muhlenbergiinae
(Poaceae: Chloridoideae: Cynodonteae) based on platid and nuclear DNA
sequences. American J. Bot. 97: 1532-1554.
Pirie, M. D. [et al. 2008], Humphreys, A. M.,
Galley, C., Barker, N. P., Verboom, G. A., Orlovich, D., Draffin, S. J., Lloyd,
K., Baeza, C. M., Negritto, M., Ruiz, E., Cota-Sánchez, J. H., Reimer, E.,
& Linder, H. P. 2008. A novel supermatrix approach improves resolution of
phylogenetic relationships in a comprehenesive sample of danthonioid grasses. Molec. Phyl. Evol. 48: 48: 1106-1119.
Pirie, M. D. [et al. 2009],
Humphreys, A. M., Barker, N. P., & Linder, H. P. 2009. Reticulation,
data combination, and inferring evolutionary history: An example from
Danthonioideae (Poaceae). Syst. Biol. 58: 612-628.
Poinar, G. O. Jr. 2004. Programinis burmitis
gen. et sp. nov., and P. laminatus sp. nov., early Cretaceous
grass-like monocots in Burmese amber. Australian Syst. Bot. 17: 497-504.
Prasad, V. [et al. 2005], Strömberg, C. A. E.,
Alimohammadian, H., & Sahni, H. 2005. Dinosaur coprolites and the early
evolution of grasses and grazers. Science 310: 1177-1180.
Preston, J.
C., & Kellogg, E. A. 2006. Reconstructing the evolutionary history of
paralogous APETALA1/FRUITFULL-like genes in grasses (Poaceae). Genetics
174: 421-437.
Preston, J.
C., & Kellogg, E. A. 2008. Discrete developmental roles for temperate
cereal grassVERNALIZATION1/FRUITFULL-like genes in flowering
competency and the transition to flowering. Plant Physiol. 146:
265-276.
Quintinar, A. [et al. 2007],
Castroviejo, S., & Catalán, P. 2007. Phylogeny of the tribe Aveneae
(Pooideae, Poaceae) inferred from plastid trnT-F and nucear ITS sequences.
American J. Bot. 94: 1554-1569.
Quintinar, A. [et al. 2010],
Catalán, P., & Castroviejo, S. 2010. A review of the systematics and
phylogenetics of the Koeleriinae (Poaceae: Poeae). Pp. 539-556, in Seberg, O.,
Petersen, G., Barfod, A. S., & Davis, J. I. (eds), Diversity, Phylogeny,
and Evolution in the Monocotyledons. Aarhus University
Press, Århus.
Reinheimer, R., & Kellogg, E. A. 2009.
Evolution of AGL6-like MADS box genes in grasses (Poaceae): Ovule
expression is ancient and palea expression is new. Plant Cell 21:
2591-2605.
Reinheimer, R., & Vegetti, A. C. 2008.
Inflorescence diversity and evolution in the PCK clade (Poaceae: Panicoideae:
Paniceae). Plant Syst. Evol. 275: 133-167.
Reinheimer, R. [et al. 2009], Zuloaga, F. O.,
Vegetti, A. C., & Pozner, R. 2009. Diversification of inflorescence
development in the PCK clade (Poaceae: Panicoideae: Paniceae). American J.
Bot. 96: 549-564.
Retallack, G. J. 2001. Cenozoic expansion of
grasslands and climate cooling. J. Geol. 109: 407-426.
Rhodes, D., & Hanson, A. D. 1993. Quaternary
ammonium and tertiary sulfonium compounds in higher plants. Ann. Rev. Plant
Physiol. Plant Mol. Biol. 44: 357-384.
Ripley, B. [et al. 2010], Frole, K., & Gilbert,
M. 2010. Difference in drought sensitivities and photosynthetic limitations
between co-occuring C3 and C4 (NADP-ME) panicoid grasses.
Ann. Bot. 105: 493-503.
Romashchenko, K. [et al. 2007], Peterson, P. M.,
Garcia-Jacas, N., Soreng, R. J., & Alfonso, S. 2007. The phylogeny of
Stipeae (Poaceae) based on nuclar DNA (ITS) sequence data. Pp. 11-12, in Botany
and Plant Biology 2007 Joint Congress. Recent Topics Poster and Changes. Chicago.
Romashchenko, K. [et al.
2008], Peterson, P. M., Soreng, R. J., Garcia-Jacas, N., Futoma, O., &
Susanna, A. 2008. Molecular phylogenetic analysis of the American
Stipeae (Poaceae) resolves Jarava sensu lato polyphyletic: Evidence for
a new genus, Pappopstipa. J. Bot. Res. Inst. Texas 2: 165-192.
Romashchenko, K. [et al. 2009], Peterson, P. M.,
Soreng, R. J., Garcia-Jacas, N., & Susanna, A. 2009. A phylogeny of Stipeae
(Poaceae: Pooideae) based on plastid and nuclear DNA sequence data. P. 189 in Botany
and Mycology 2009. Snowbird, Utah
July 25-29. Abstract Book.
Romashchenko, K. [et al. 2010], Peterson, P. M.,
Soreng, R. J., Garcia-Jacas, N., & Susanna, A. 2009. Phylogenetics of
Stipeae (Poaceae: Pooideae) based on plastid and nuclear DNA sequences. Pp.
511-537, in Seberg, O., Petersen, G., Barfod, A. S., & Davis, J. I. (eds), Diversity,
Phylogeny, and Evolution in the Monocotyledons. Aarhus University
Press, Århus.
Rondeau, R., Rouch, C., & Besnard, G. 2005.
NADP-malate dehydrogenase gene evolution in Andropogoneae (Poaceae): Gene
duplication folllowed by sub-functionalization. Ann. Bot. 96: 1307-1314.
Rua, G. H. [et al. 2010],
Speranza, P. R., Vaio, M., & Arakaki, M. 2010. A
phylogenetic analysis of the genus Paspalum (Poaceae) based on cpDNA and
morphology. Plant Syst. Evol. 288: 227-243.
Rudall, P. J. [et al. 2005a], Stuppy, W., Cunniff,
J., Kellogg, E. A., & Briggs, B. G. 2005a. Evolution of reproductive
structures in grasses (Poaceae) inferred by sister-group comparison with their
putative closest living relatives, Ecdeiocoleaceae. American J. Bot. 92:
1432-1443.
Saarela, J. M., & Graham, S. W. 2010. Inference
of phylogenetic relationships among subfamilies of grasses (Poaceae: Poales)
using meso-scale exemplar-based sampling of the plastid genome. Botany
88: 65-84.
Saarela, J. M. [et al. 2010], Liu, Q., Peterson, P.
M., Soreng, R. J., & Paszko, B. 2010. Phylogenetic of the grass 'Aveneae-type
plastid DNA clade' (Poaceae: Pooideae, Poeae) based on plastid and ribosomal
DNA sequence data. Pp. 557-587, in Seberg, O., Petersen, G., Barfod, A. S.,
& Davis, J. I. (eds), Diversity, Phylogeny, and Evolution in the
Monocotyledons. Aarhus
University Press, Århus.
Sabelli, P. A., & Larkins, B. A. 2009. The
development of endosperm in grasses. Plant Physiol. 149: 14-26.
Sage, R. F., & Kubien, D. S. 2003. Quo vadis
C(4)? An ecophysiological perspective on global change and the future of C(4)
plants. Photosynth. Res. 77: 209-225.
Sage, R. F. [et al. 1999], Li,
M., & Monson, R. K. 1999. The taxonomic distribution of C4
photosynthesis. Pp. 551-584, in Sage, R. F., & Monson, R. K. (eds), C4
Plant Biology. Academic Press, San
Diego.
Sage, R. F. [et al. 2011], Christin, P.-A., &
Edwards, E. J. 20. The C4 lineages of planet earth. J.
Experiment. Bot. 62: 3155-3169.
Salamin, N., & Davies, T. J. 2004. Using
supertrees to investigate species richness in grasses and flowering plants. Pp.
461-486, in Bininda-Emonds, O. R. P. (ed.), Phylogenetic Supertrees:
Combining Information to Reveal the Tree of Life. Kluwer Academic, Dordrecht.
Salariato, D. L. [et al. 2010], Zuloaga, F. O.,
Giussani, L. M., & Morrone, O,. 2010. Molecular phylogeny of the subtribe
Melinidinae (Poaceae: Panicoidae: Paniceae) and evolutionary trends in the
homogenization of inflorescences. Mol. Phyl. Evol. 56: 355-369.
Sánchez-Ken, J. G., & Clark, L. G. 2007 [2008].
Phylogenetic relationships within the Centothecoideae + Panicoideae clade
(Poaceae) based on ndhF and rpl16 intron sequences and structural
data. Pp. 487-502, in Columbus, J. T., Friar, E. A., Porter, J. M., Prince, L.
M., & Simpson, M. G. (eds), Monocots: Comparative Biology and Evolution.
Poales.. Rancho Santa Ana Botanical Garden, Claremont, Ca.
[Aliso 23: 487-502.]
Sánchez-Ken, J. G., &
Clark, L. G. 2010. Phylogeny and a new tribal classification of the
Panicoideae s.l. (Poaceae) based on plastid and nuclear sequence data and
structural data. American J. Bot. 97: 1732-1748.
Sánchez-Ken, J. G. [et al. 2007], Clark, L. G.,
Kellogg, E. A., & Kay, E. E. 2007. Reinstatement and emendation of
subfamily Micrairoideae (Poaceae). Syst. Bot. 32: 71-80.
Sandve, S. R., & Fjellheim, S. 2010. Did gene
family expansions during the Eocene-Oligocene boundary climate cooling play a
part in Pooideae adaptation to cool climates? Molec. Evol. 10:
2075-2088.
Sandve, S. R. [et al. 2008], Rudi, H., Asp, T.,
& Rognli, O. A. 2008. Tracking the evolution of a cold stress associated
gene family in cold tolerant grasses. BMC Evol. Biol. 8: 245.
doi:10.1186/1471-2148-8-245
Saski, C. [et al. 2007], Lee, S. B., Fjellhem, S.,
Guda, C., Jansen, R. K., Luo, H., Tomkins, J., Rognli, O. A., Daniell, H.,
& Clarke, J. L. 2007. Complete chloroplast gemome sequences of Hordeum vulgare,
Sorghum bicolor and Agrostis stolonifera, and comparative
analyses with other grass genomes. Theor. Appl. Genetics 115: 571-590.
Schardl, C. L. 2010. The Epichloae, symbionts of
the grass subfamily Poöideae. Ann. Missouri
Bot. Gard. 97: 646-665.
Schlueter, J. A. [et al. 2004], Dixon, P., Granger,
C., Grant, D., Clark, L., Doyle, J. J., & Schoenmaker, R. C. 2004. Mining
EST databases to resolve evolutionary events in major crop species. Genome
47: 868-876.
Schneider, J. [et al. 2009], Döring, E., Hilu, K.
W., & Röser, M. 2009. Phylogenetic structure of the grass subfamily
Pooideae based on comparison matK gene-3'trnK exon and nuclear
ITS sequences. Taxon 58: 405-424.
Schneider, J. [et al. 2011], Winterfeld, G.,
Hoffmann, M. H., & Röser, M. 2011. Duthieeae, a new tribe of grasses
(Poaceae) identified among the early diverging lineages of subfamily Pooideae:
Molecular phylogenetics, morphological delineation, cytogenetics and
biogeopgraphy. Syst. Biodivers. 9: 27-44.
Schullehner, K. [et al. 2008], Dick, R., Vitzthum,
F., Schwab, W., Brandt, W., Frey, M., & Gierl, A. 2008. Benzoxazinoid
biosynthesis in dicot plants. Phytochem. 69: 2668-2677.
Sede, S. M. [et al. 2008],
Morrone, O., Giussani, L. M., & Zuloaga, F. O. 2008. Phylogenetic
studies in Paniceae (Poaceae): A realignment of section Lorea of Panicum.
Syst. Bot. 33: 284-300.
Sede, S. M. [et al. 2009a], Morrone, O., Aliscioni,
S. S., Giussani, L. M., & Zuloaga, F. O. 2009a. Oncorachis and Sclerochlamys,
two new segregated genera from Streptostachys (Poaceae, Panicoideae,
Paniceae): A revision based on molecular, morphological and anatomical
characters. Taxon 58: 365-374.
Sede, S. M. [et al. 2009b],
Zuloaga, F. O., & Morrone, O. 2009b. Phylogenetic studies in the
Paniceae (Poaceae-Panicoideae); Ocellochloa, a new genus from the New World. Syst. Bot. 34: 684-6932.
Sendulsky, T. [et al. 1986],
Filguerias, T. S., & Burman, A. G. 1986. Fruits, embryos and
seedlings. Pp. 31-36, in Soderstrom, T. R., Hilu, K. W., Campbell, C. S., &
Barkworth, M. E. (eds), Grass Systematics and Evolution, Smithsonian
Institute Press, Smithsonian Institution.
Simon, B. K. 2007 [2008]. Grass phylogeny and
classification: Conflict of morphology and molecules. Pp. 259-266, in Columbus,
J. T., Friar, E. A., Porter, J. M., Prince, L. M., & Simpson, M. G. (eds), Monocots:
Comparative Biology and Evolution. Poales.. Rancho Santa Ana
Botanical Garden, Claremont, Ca. [Aliso 23: 259-266.]
Sindhu, A. [et al. 2008],
Chintamanani, S., Brandt, A. S., Zanis, M., Scofield, S. R., & Johal, G. S.
2008. A guardian of grasses: Specific origin and conservation of a unique
disease-resistance gene in the grass lineage. Proc. National Acad. Sci.
U.S.A. 105: 1762-1767.
Smith, S. A., & Donoghue, M. J. 2008. Rates of
molecular evolution are linked to life history in flowering plants. Science
322: 86-89.
Smith, S. A. [et al. 2011],
Beaulieu, J. M., Stamatkis, A., & Donoghue, M. J. 2011. Understanding
angiosperm diversification using small and large phylogenetic trees. American
J. Bot. 98:
Smith, S. Y. [et al. 2010], Collinson, M. E.,
Rudall, P. J., & Simpson, D. A. 2010. Cretaceous and Paleogene fossil
record of Poales: Review and current research. Pp. 333-356, in Seberg, O.,
Petersen, G., Barfod, A. S., & Davis, J. I. (eds), Diversity, Phylogeny,
and Evolution in the Monocotyledons. Aarhus University
Press, Århus.
Soreng, R. J., & Davis, J. I. 1998.
Phylogenetics and character evolution in the grass family (Poaceae):
Simultaneous analysis of morphological and chloroplast DNA restriction site
character sets. Bot Rev. 64: 1-85.
Soreng, R. J., & Davis, J. I. 2000.
Phylogenetic structure in Poaceae subfamily Pooideae as inferred from molecular
and morphological characters: Misclassification versus reticulation. Pp. 61-74,
in Jacobs, S. W. L., & Everett, J. (eds), Grasses: Systematics and
Evolution. CSIRO, Melbourne.
Soreng, R. J. [et al. 2003], Peterson, P. M.,
Davidse, G., Judziewicz, E. J., Zuloaga, F. O., Filgueiras, T. S., &
Morrone, O. 2003. Catalogue of New World
grasses (Poaceae): IV. Subfamily Pooideae. Contr. U. S. National Herb.
48: 1-730.
Soreng, R. J. [et al. 2007], Davis, J. I., &
Voionomaa, M. A. 2007. A phylogenetic analysis of Poaceae tribe Poeae sensu
lato based on morphological characters and sequence data from three
plastid-encoded genes: Evidence for reticulation, and a new classification of
the tribe. Kew Bull. 62:
425-454.
Soreng, r. J. [et al. 2010], Bull, R. D., &
Gillespie, L. J. 2010. Phylogeny and reticulation in Poa based on
plastid trnTLF and nrITS sequences with attention to diploids. Pp. 619-643,
in Seberg, O., Petersen, G., Barfod, A. S., & Davis, J. I. (eds), Diversity,
Phylogeny, and Evolution in the Monocotyledons. Aarhus University
Press, Århus.
Strömberg, C. A. E. 2005. Decoupled taxonomic
radiation and ecological expansion of open-habitat grasses in the Cenozoic of
North America. Proc. National Acad. Sci. U.S.A. 102: 11980-11984.
Strömberg, C. A. E., & McInerney, F. A. 2011.
The Neogene transition from C3 to C4 grasslands in North America: Assemblage analysis of fossil phytoliths. Paleobiol.
37: 50-71.
Sungkaew, S. [et al. 2009], Stapleton, C. M. A.,
Salamin, N., & Hodkinson, T. R. 2009. Non-monophyly of woody bamboos
(Bambuseae, Poaceae): A multi-gene region phylogenetic analysis of Bambusoideae
s.s. J. Plant Res. 122: 95-108.
Sungkaew, S. [et al. 2010], Stapleton, C. M. A.,
& Hodkinson, T. R. 2010. Phylogentics of Dendrocalamus (Poaceae:
Bambusoideae, Bambuseae, Bambusinae) and its related genera based on combined
multi-gene analyses. Pp. 497-510, in Seberg, O., Petersen, G., Barfod, A. S.,
& Davis, J. I. (eds), Diversity, Phylogeny, and Evolution in the
Monocotyledons. Aarhus
University Press, Århus.
Tang, L. [et al. 2010], Zou, X.-H., Achoundong, G.,
Potgieter, C., Second, G., Zhang, D.-Y., & Ge, S. 2010. Phylogeny and
biogeography of the rice tribe (Oryzeae): Evidence from combined analysis of 20
chloroplast fragments. Mol. Phyl. Evol. 54: 266-277.
Taylor, D. W. [et al. 2010], Zinniker, D.,
McCorkle, E., Hu, S., Barbanti, S. M., & Moldowan, J. M. 2010. Tracking
sngiosperm molecular fossils: New results from analysis of basal living clades.
P. 73, in Botany 2010. July 31 - August 4, Providence, Rhode Island.
Scientific Abstracts.
Thorne, R. F. 2007. An updated classification of
the class Magnolipsida ("Angiospermae"). Bot. Review 73: 67-182.
["with many nomenclatural additions by James L. Reveal."; for online
version, see http://www.rsabg.org/content/view/50/144/]
Tipple, B. J., & Pagani, M. 2007. The early
origins of terrestrial C4 photosynthesis. Ann. Rev. Earth Planet.
Sci. 35: 435-461.
Tremblay, R. L. [et al. 2005], Ackerman, J. D.,
Zimmerman, J. K., & Calvo, R. N. 2005. Variation in sexual reproduction in
orchids and its evolutionary consequences: A spasmodic journey to
diversification. Biol. J. Linnean Soc. 84: 1-54.
Tyrrell, C. D. [et al. 2009], Triplett, J. K.,
Santos-Gonçalves, A. P., Londono, X., & Clark, L. G. 2009. Phylogeny,
morphological evolution, and generic classification within Arthrostylidiinae
(Poaceae: Bambusoideae: Bambuseae). P. 183, in Botany and Mycology 2009.
Snowbird, Utah
July 25-29. Abstract Book.
Vester, von H. 1940. Die Areale und Arealtypen der
Angiospermen-Familien. Bot. Arkiv 41: 203-275, 295-356.
Vicentini, A. [et al. 2008], Barber, J. C.,
Aliscioni, S. S., Giussani, L. M., & Kellogg, E. A. 2008. The age of the
grasses and clusters of origins of C4 photosynthesis. Global
Change Biol. 14: 1-15.
Wang, D. [et al. 2008], Portis, A. R. Jr., Moose,
S. P., & Long, S. P. 2008. Cool C4 photosynthesis: Pyruvate Pi
dikinase expression and activity corresponds to the exceptional cold tolerance
of carbon assimilation in Miscanthus x giganteus. Plant Physiol.
148: 557-567.
Watson, L., & Dallwitz, M. 1992b onwards. Grass
Genera of the World: Descriptions, Illustrations, Identification, and
Information Retrieval.... http://delta-intkey.com
Wedin, D. A. 1995. Species, nitrogen, and grassland
dynamics: The constraints of stuff. Pp. 253-262, in Jones, C. G., & Lawton, J. H. (eds), Linking
Species and Ecosystems. Chapman and Hall, New York.
Wikström, N. [et al. 2001], Savolainen, V., &
Chase, M. W. 2001. Evolution of the angiosperms: Calibrating the family tree. Proc.
Roy. Soc. B, 268: 2211-2220.
Winterfeld, G. 2006. Molekular-cytogenetische
Untersuchungen an Hafergräsern (Aveneae) und anderer Poaceae. Stapfia
86: 1-170.
Yang, H.-Q. [et al. 2008a],
Wang, H., & Li, D.-Z. 2008a. Comparative morphology of the
foliage leaf epidermis, with emphasis on papillae characters, in key taxa of
woody bamboos of the Asian tropics (Poaceae: Bambusoideae). Bot. J. Linnean
Soc. 156: 411-423.
Yang, H.-Q. [et al. 2008b], Yang, J.-B., Peng,
Z.-H., Gao, J., Yang, Y.-M., Peng, S., & Li, D.-Z. 2008b. A molecular
phylogenetic and fruit evolutionary analysis of the major groups of the paleotropical
woody bamboos (Gramineae: Bambusoideae) based on nuclear ITS, GBSSI and plastid
trnL-F DNA sequences. Molec.
Phyl. Evol. 48: 809-824.
Yang, J. B. [et al. 2010],
Yang, H.-Q., Li, D.-Z., Wong, K.-M., & Yang, Y.-M. 2010. Phylogeny
of Bambusa and its allies (Poaceae: Bambusoideae) inferred from nuclear GBSSI
gene and plastid psbA-trnH, rpl32-trnL and rps16
intron DNA sequences. Taxon 59: 1102-1110.
Zachos, J. C. [et al. 2008],
Dickens, G. R., & Zeebe, R. E. 2008. An early Cenozoic perspective
on greenhouse warming and carbon-cycle dynamics. Nature 451: 279-283.
Zeng, C.-X. [et al. 2010],
Zhang, Y.-X., Triplett, J. K., Yang, J.-B., & Li, D.-Z. 2010. Large
multi-locus plastid phylogeny of the tribe Arundinarieae (Poaceae:
Bambusoideae) reveals ten major lineages and low rate of molecular divergence. Mol.
Phyl. Evol. 56: 821-839.
Zhang, W.[eiping], & Clark, L. G. 2000.
Phylogeny and classification of the Bambusoideae (Poaceae). Pp. 35-42, in
Jacobs, S. W. L., & Everett, J. (eds), Grasses: Systematics and
Evolution. CSIRO, Melbourne.
Zou, X.-H. [et al. 2008],
Zhang, F.-M., Zhang, J.-G., Zang, L.-L., Tang, L., Wang, J., Sang, T., &
Ge, S. 2008. Analysis of 142 genes resolves the rapid
diversification of the rice genus. Genome Biol. 9:R49. [htttp://genomebiology.com/2008/9/3/R49]
Zuloaga, F. O. [et al. 2000], Morrone, O., &
Giussani, L. M. 2000. A cladistic analysis of the Paniceae: A preliminary
approach. Pp. 123-135, in Jacobs, S. W. L., & Everett, J. (eds), Grasses:
Systematics and Evolution. CSIRO, Melbourne.
Zuloaga, F. O. [et al. 2007], Giussani, L. M.,
& Morrone, O. 2007. Hopia, a new monotypic genus segregated from Panicum
(Poaceae). Taxon 56: 145-156.
Zuloaga, F. O. [et al. 2010], Scataglini, M. A.,
& Morrone, O. 2010. A phylogenetic evaluation of Panicum sects. Agrostoidea,
Megista, Prionotia and Tenera (Panicoideae, Poaceae): Two
new genera, Stephostachys and Sorengia. Taxon 59:
1535-1546.
